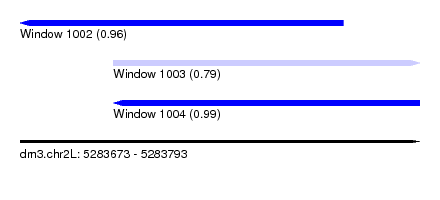
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,283,673 – 5,283,793 |
| Length | 120 |
| Max. P | 0.994346 |

| Location | 5,283,673 – 5,283,770 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.73 |
| Shannon entropy | 0.27535 |
| G+C content | 0.45414 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.69 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
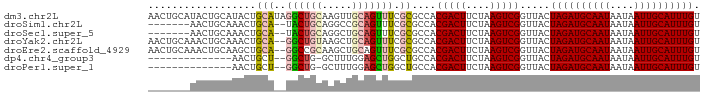
>dm3.chr2L 5283673 97 - 23011544 GGCUGCAAGUUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAA-CGAG----------- ((((((.....))))))(((((((((((((....)))))(..((.(((((((((....)))))))))))..)..............)))))..-))).----------- ( -30.40, z-score = -3.23, R) >droSim1.chr2L 5085814 90 - 22036055 -------GGCCGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAA-CGAG----------- -------..........(((((((((((((....)))))(..((.(((((((((....)))))))))))..)..............)))))..-))).----------- ( -24.00, z-score = -2.20, R) >droSec1.super_5 3362700 90 - 5866729 -------GGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAA-CGAG----------- -------..........(((((((((((((....)))))(..((.(((((((((....)))))))))))..)..............)))))..-))).----------- ( -24.00, z-score = -2.14, R) >droYak2.chr2L 8409896 98 + 22324452 GGCUGUAAGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAACCGAG----------- ((((((.....))))))(((((((((((((....)))))(..((.(((((((((....)))))))))))..)..............)))))...))).----------- ( -27.80, z-score = -2.35, R) >droEre2.scaffold_4929 5363961 97 - 26641161 GGCCGCAAGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAA-CGAG----------- .(((....)..))....(((((((((((((....)))))(..((.(((((((((....)))))))))))..)..............)))))..-))).----------- ( -25.90, z-score = -1.64, R) >droAna3.scaffold_12916 6302750 96 + 16180835 -------------GUUUUCGUGGCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAACAACGACGACGUCGAG -------------......(((((.(((((....))))))))))((((((((((....))))))))))................(((((((...........))))))) ( -25.90, z-score = -2.40, R) >dp4.chr4_group3 7087206 88 - 11692001 ---------UUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCCAA-CGUG----------- ---------.....(.(((((....(((((....)))))(..((.(((((((((....)))))))))))..)...............))))).-)...----------- ( -22.20, z-score = -1.71, R) >droPer1.super_1 8541062 88 - 10282868 ---------UUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCCAA-CGUG----------- ---------.....(.(((((....(((((....)))))(..((.(((((((((....)))))))))))..)...............))))).-)...----------- ( -22.20, z-score = -1.71, R) >consensus ________GCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGUCCCACUUUACUUUCACUUGGCGAA_CGAG___________ ....................((((((((((....)))))(..((.(((((((((....)))))))))))..)..............))))).................. (-20.39 = -20.69 + 0.30)


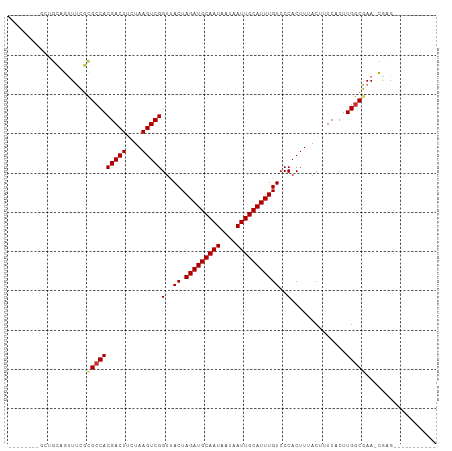
| Location | 5,283,701 – 5,283,793 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Shannon entropy | 0.33064 |
| G+C content | 0.44430 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -16.07 |
| Energy contribution | -17.34 |
| Covariance contribution | 1.27 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5283701 92 + 23011544 ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAACUUGCAGCCUAUGCAGUAUGCAGUAUGCAGUU ....(((((((....)))))))...((...(((((....)))))..))(((..(((((((((.(((.....)))))).)))))).))).... ( -27.60, z-score = -1.67, R) >droSim1.chr2L 5085842 83 + 22036055 ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCGGCCUGCAGUA--UGCAGUUUGCAGUU------- ....(((((((....)))))))...((..((((((....))))).)..))..(((((((..((((....--.))))..)))))))------- ( -28.60, z-score = -2.64, R) >droSec1.super_5 3362728 83 + 5866729 ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAGCCUGCAGUA--UGCAGUUUGCAGUU------- ....(((((((....)))))))...((..((((((....))))).)..))..((((((((.((((....--.)))).))))))))------- ( -29.30, z-score = -3.19, R) >droYak2.chr2L 8409925 90 - 22324452 ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAGCUUACAGCC--UGCAGUUUGCAGUUUGCAGUU .....(((((.....(((((.....((..((((((....))))).)..)).((((((((((.....).)--))))))))))))))))))... ( -28.40, z-score = -2.02, R) >droEre2.scaffold_4929 5363989 90 + 26641161 ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAGCUUGCGGCC--UGCAGCUUGCAGUUUGCAGUU ....(((((((....)))))))...((...(((((....)))))..))((.(((((((((((.((....--.))))).)))))))))).... ( -30.50, z-score = -1.60, R) >dp4.chr4_group3 7087234 75 + 11692001 ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCAGCCAGCUCCAAAGC-CAGCC--AGCAGUU-------------- ....(((((((....)))))))...((...(((((....)))))((((.....(((....)))-..)))--)))....-------------- ( -16.50, z-score = -1.27, R) >droPer1.super_1 8541090 75 + 10282868 ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCAGCCAGCUCCAAAGC-CAGCC--AGCAGUU-------------- ....(((((((....)))))))...((...(((((....)))))((((.....(((....)))-..)))--)))....-------------- ( -16.50, z-score = -1.27, R) >consensus ACAAAUGCAAUUAUUAUUGCAUCUAGUAACCGACUUAGAAGUCGUGGCGCGAAACUGCAGCUUGCAGCC__UGCAGUUUGCAGUU_______ ....(((((((....)))))))...((...(((((....)))))..))((.(((((((..............)))))))))........... (-16.07 = -17.34 + 1.27)

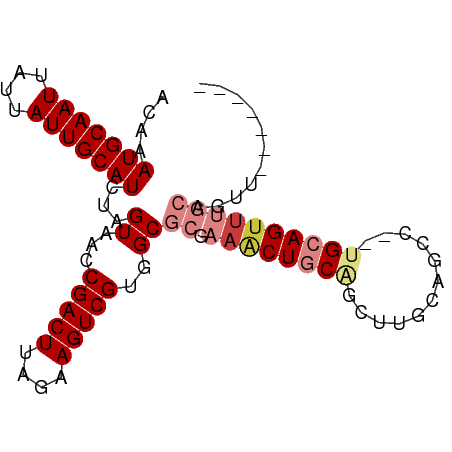
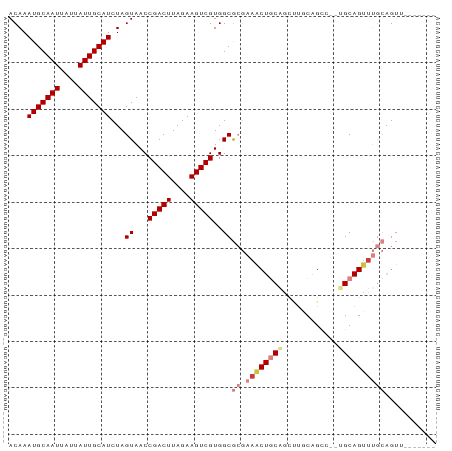
| Location | 5,283,701 – 5,283,793 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Shannon entropy | 0.33064 |
| G+C content | 0.44430 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -19.37 |
| Energy contribution | -19.89 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
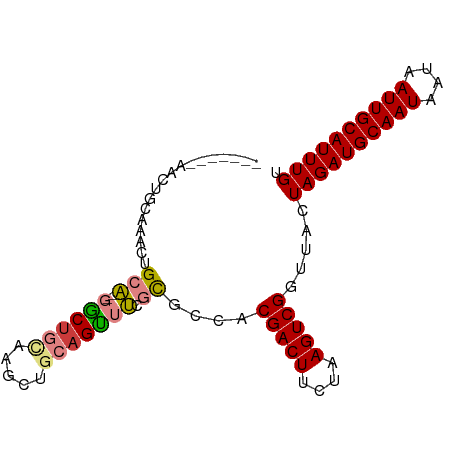
>dm3.chr2L 5283701 92 - 23011544 AACUGCAUACUGCAUACUGCAUAGGCUGCAAGUUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU ....((..((((((.((((((.....))).)))))))))...))....(((((....))))).....((((((((((....)))))))))). ( -28.90, z-score = -2.19, R) >droSim1.chr2L 5085842 83 - 22036055 -------AACUGCAAACUGCA--UACUGCAGGCCGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU -------((((((...((((.--....))))...))))))........(((((....))))).....((((((((((....)))))))))). ( -27.50, z-score = -3.09, R) >droSec1.super_5 3362728 83 - 5866729 -------AACUGCAAACUGCA--UACUGCAGGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU -------(((((((..((((.--....))))..)))))))........(((((....))))).....((((((((((....)))))))))). ( -30.50, z-score = -4.09, R) >droYak2.chr2L 8409925 90 + 22324452 AACUGCAAACUGCAAACUGCA--GGCUGUAAGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU .......(((.((((((((((--(........))))))))).))....(((((....))))))))..((((((((((....)))))))))). ( -29.50, z-score = -2.59, R) >droEre2.scaffold_4929 5363989 90 - 26641161 AACUGCAAACUGCAAGCUGCA--GGCCGCAAGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU ....((((((((((.(((((.--....)).))))))))))).))....(((((....))))).....((((((((((....)))))))))). ( -32.80, z-score = -2.96, R) >dp4.chr4_group3 7087234 75 - 11692001 --------------AACUGCU--GGCUG-GCUUUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU --------------....((.--(((..-(((....)))..)))))..(((((....))))).....((((((((((....)))))))))). ( -26.20, z-score = -3.09, R) >droPer1.super_1 8541090 75 - 10282868 --------------AACUGCU--GGCUG-GCUUUGGAGCUGGCUGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU --------------....((.--(((..-(((....)))..)))))..(((((....))))).....((((((((((....)))))))))). ( -26.20, z-score = -3.09, R) >consensus _______AACUGCAAACUGCA__GGCUGCAAGCUGCAGUUUCGCGCCACGACUUCUAAGUCGGUUACUAGAUGCAAUAAUAAUUGCAUUUGU ..................((...((((((.....))))))....))..(((((....))))).....((((((((((....)))))))))). (-19.37 = -19.89 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:40 2011