| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,176,129 – 15,176,248 |
| Length | 119 |
| Max. P | 0.917351 |

| Location | 15,176,129 – 15,176,248 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.61744 |
| G+C content | 0.41394 |
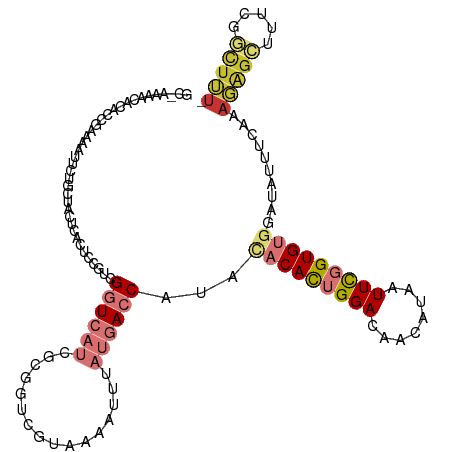
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -10.16 |
| Energy contribution | -11.62 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
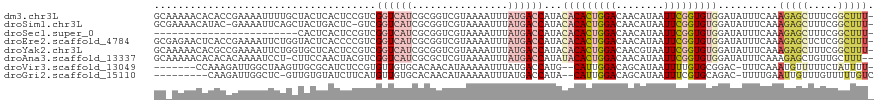
>dm3.chr3L 15176129 119 + 24543557 GCAAAAACACACCGAAAAUUUUGCUACUCACUCCGUCGGUCAUCGCGGUCGUAAAAUUUAUGACCAUACACACUGGACAACAUAAUUCGGUGUGGAUAUUUCAAAGAGCUUUCGGCUUU- ((((((............))))))..........(((((...((..(((((((.....)))))))...(((((((((........)))))))))...........))....)))))...- ( -29.70, z-score = -2.45, R) >droSim1.chr3L 14510458 117 + 22553184 GCGAAAACAUAC-GAAAAUUCAGCUACUGACUC-GUCGGUCAUCGCGGUCGUAAAAUUUAUGACCAUACACACUGGACAACAUAAUUCGGUGUGGAUAUUUCAAAGAGCUUUCGGCUUU- ((((..((..((-((....((((...)))).))-))..))..))))(((((((.....)))))))...(((((((((........)))))))))..........(((((.....)))))- ( -34.60, z-score = -3.65, R) >droSec1.super_0 7288634 95 + 21120651 ------------------------CACUCACUCCGUCGGUCAUCGCGGUCGUAAAAUUUAUGACCAUACACACUGGACAACAUAAUUCGGUGUGGAUAUUUCAAAGAGCUUUCGGCUUU- ------------------------..........(((((...((..(((((((.....)))))))...(((((((((........)))))))))...........))....)))))...- ( -26.70, z-score = -2.91, R) >droEre2.scaffold_4784 17438292 119 + 25762168 GCGAGAACUCACCGAAAAUUCUGGUACUCACCCCGUCGGUCAUCGCGGUCGUAAAAUUUAUGACCAUACACACUGGACAACAUAAUUCGGUGUGGAUAUUUCAAAGAGCUCUCGGCUUU- .(((((.((((((((.......(((....)))...))))).(((..(((((((.....)))))))...(((((((((........))))))))))))........))).))))).....- ( -38.80, z-score = -3.77, R) >droYak2.chr3L 17683626 119 + 24197627 GCAAAAACACGCCGAAAAUUCUGGUGCUCACUCCGUCGGUCAUCGCGGUCGUAAAAUUUAUGACCAUACACACUGGACAACGUAAUUCGGUGUGGAUAUUUCAAAGAGCUUUCGGCUUU- ..........(((((((.((((((((((.((...)).)).))))..(((((((.....)))))))...(((((((((........)))))))))..........)))).)))))))...- ( -36.80, z-score = -3.07, R) >droAna3.scaffold_13337 5479770 117 - 23293914 GCAAAAACACACACAAAAUCCU-CUUCCAACUACGUCGGUCAUCGCGCUCGUAAAAUUUAUGACCAUAUACACUGGACAACAUAAUUCGGUGUGGAUAUUUCAAAGAGCUGUUGCUUU-- .................((((.-..............((((((................))))))....((((((((........)))))))))))).......(((((....)))))-- ( -21.29, z-score = -0.69, R) >droVir3.scaffold_13049 19971287 109 + 25233164 -------CCAAAGAUUGGCUAAGUUGCGCAUCUCCGUGUUGUGCACAACAUAAAAAUUUAUGACCAUG--CAUUGGACAGCAUAAUUUUGUGCGGAC-UUUCAAAUGUUUUUCUAUUUU- -------....(((..(((.((((((((((.....(((((((......(((((....))))).(((..--...)))))))))).....))))).)))-))......)))..))).....- ( -24.60, z-score = -0.71, R) >droGri2.scaffold_15110 4368541 107 - 24565398 ---------CAAGAUUGGCUC-GUUGUGUAUCUUCAUGUUGUGCACAACAUAAAAAUUUAUGACCAUA--CAUUGGACAGCAUAAUUUCGUGCAGAC-UUUUGAAUUGUUUGUUUUUGUC ---------(((((..(((..-(((((((((.........))))))))).....((((((((.(((..--...))).))((((......))))....-...)))))))))...))))).. ( -20.70, z-score = 0.46, R) >consensus GC_AAAACACACCGAAAAUUCUGCUACUCACUCCGUCGGUCAUCGCGGUCGUAAAAUUUAUGACCAUACACACUGGACAACAUAAUUCGGUGUGGAUAUUUCAAAGAGCUUUCGGCUUU_ .....................................((((((................))))))...(((((((((........)))))))))..........(((((.....))))). (-10.16 = -11.62 + 1.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:06 2011