
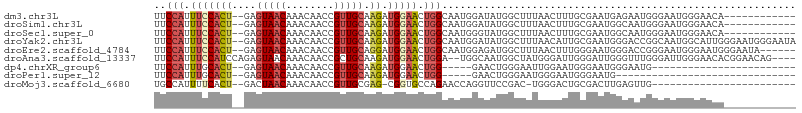
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,173,587 – 15,173,680 |
| Length | 93 |
| Max. P | 0.580144 |

| Location | 15,173,587 – 15,173,680 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Shannon entropy | 0.59736 |
| G+C content | 0.45821 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -8.10 |
| Energy contribution | -8.74 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15173587 93 + 24543557 UUCCAUUUCCACU--GAGUAACAAACAACCGUUGCAAGAUGGAACUGGCAAUGGAUAUGGCUUUAACUUUGCGAAUGAGAAUGGGAAUGGGAACA------------ (((((((((((..--.............(((((((.((......)).))))))).....((.........)).........)))))))))))...------------ ( -25.90, z-score = -2.61, R) >droSim1.chr3L 14507937 93 + 22553184 UUCCAUUUCCACU--GAGUAACAAACAACCGUUGCAAGAUGGAACUGGCAAUGGAUAUGGCUUUAACUUUGCGAAUGGCAAUGGGAAUGGGAACA------------ (((((((((((..--.............(((((((.((......)).)))))))..............((((.....)))))))))))))))...------------ ( -27.90, z-score = -2.80, R) >droSec1.super_0 7286146 93 + 21120651 UUCCAUUUCCACU--GAGUAACAAACAACCGUUGCAAGAUGGAACUGGCAAUGGGUAUGGCUUUAACUUUGCGAAUGGCAAUGGGAAUGGGAACA------------ (((((((((((..--((((.........(((((((.((......)).))))))).....)))).....((((.....)))))))))))))))...------------ ( -28.64, z-score = -2.53, R) >droYak2.chr3L 17680982 105 + 24197627 UUCCAUUUCCACU--GAGUAACAAACAACCGUUGCAAGAUGGAACUGGCAAUGGAUAUGGCUUUAACAUUGCGAAUGGGACCGGCAAUGGCAUUGGGAAUGGGAAUA (((((((((((.(--(............(((((((.((......)).)))))))............((((((...........)))))).)).)))))))))))... ( -33.00, z-score = -2.75, R) >droEre2.scaffold_4784 17435146 99 + 25762168 UUCCAUUUCCACU--GAGUAACAAACAACCGUUGCAGGAUGGAACUGGCAAUGGAGAUGGCUUUAACUUUGGGAAUGGGACCGGGAAUGGGAAUGGGAAUA------ (((((((((((((--(.((..((.....(((((((((.......)).)))))))......((........))...))..)))))...)))))))))))...------ ( -29.00, z-score = -2.01, R) >droAna3.scaffold_13337 5477404 101 - 23293914 UUCCAUUUCCAUCCAGAGUAACAAACAACCGCUGCAAGAUGGAACUGGA--UGGCAAUGGCUAUGGGAUUGGGAUUGGGUUUGGGAUUGGGAACACGGAACAG---- (((((.((((((((((.((.....))..((((.....).)))..)))))--((((....))))))))).)))))....((((.(...((....))).))))..---- ( -25.00, z-score = -0.01, R) >dp4.chrXR_group6 12750189 76 - 13314419 UUCCAUUUGCACU--GAGUAACAAACAACCGUUGCAAGAUGGAACUGG-----GAACUGGGAAUUGGAAUGGGAAUGGGAAUG------------------------ ((((((((.((((--.(((..(......(((((....))))).....)-----..))).))........)).))))))))...------------------------ ( -17.00, z-score = -1.26, R) >droPer1.super_12 1168222 70 - 2414086 UUCCAUUUGCACU--GAGUAACAAACAACCGUUGCAAGAUGGAACUGG-----GAACUGGGAAUGGGAAUGGGAAUG------------------------------ ((((((((.((((--.(((..(......(((((....))))).....)-----..))).))..)).))))))))...------------------------------ ( -18.90, z-score = -2.42, R) >droMoj3.scaffold_6680 7668619 79 + 24764193 UGCCAUUUUCACU--GACUAACAAACAACCGUUGCGAG-CGGUGCCAGAACCAGGUUCCGAC-UGGGACUGCGACUUGAGUUG------------------------ .............--((((........(((((.....)-))))((.((..((((.......)-)))..))))......)))).------------------------ ( -17.90, z-score = 0.48, R) >consensus UUCCAUUUCCACU__GAGUAACAAACAACCGUUGCAAGAUGGAACUGGCAAUGGAUAUGGCUUUAACAUUGCGAAUGGGAAUGGGAAUGGGAACA____________ ..(((.(((((((....(((((........))))).)).))))).)))........................................................... ( -8.10 = -8.74 + 0.65)
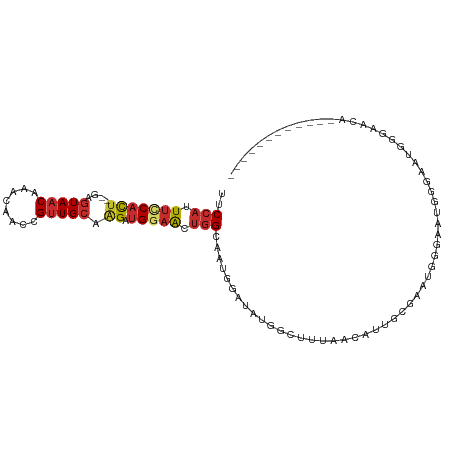
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:05 2011