| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,127,642 – 15,127,785 |
| Length | 143 |
| Max. P | 0.984153 |

| Location | 15,127,642 – 15,127,785 |
|---|---|
| Length | 143 |
| Sequences | 11 |
| Columns | 145 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.51251 |
| G+C content | 0.51805 |
| Mean single sequence MFE | -46.06 |
| Consensus MFE | -22.57 |
| Energy contribution | -21.60 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.72 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
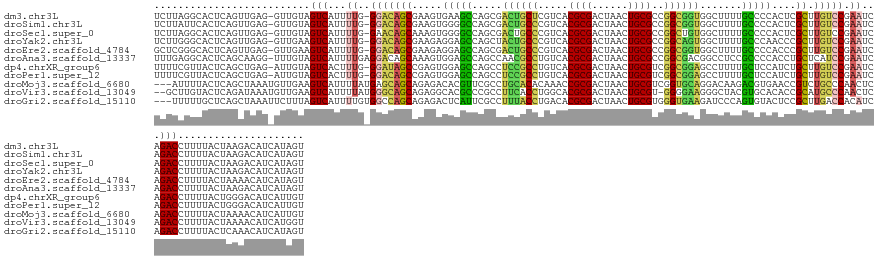
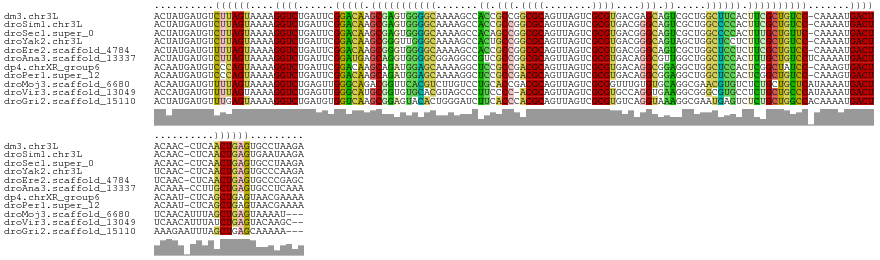
>dm3.chr3L 15127642 143 + 24543557 UCUUAGGCACUCAGUUGAG-GUUGUAGUCAUUUUG-GGACAGCGAAGUGAAGCCAGCGACUGCUCGUCACGCGACUAACUGCGCCGGCGGUGGCUUUUGCCCCACUCGCUUGUCCGAAUCAGACCUUUUACUAAGACAUCAUAGU ((((((((.(((....)))-))....(((...((.-(((((((((.(((..((.(((.(((((.((...((((......)))).))))))).)))...))..))))))).))))).))...)))......))))))......... ( -49.40, z-score = -1.60, R) >droSim1.chr3L 14459429 143 + 22553184 UCUUAUUCACUCAGUUGAG-GUUGUAGUCAUUUUG-GGACAGCGAAGUGGGGCCAGCGACUGCCCGUCACGCGACUAACUGCGCCGGCGGUGGCUUUUGCCCCACUCGCUUGUCCGAAUCAGACCUUUUACUAAGACAUCAUAGU .........((.(((.(((-(((.........((.-(((((((((.(((((((.(((.((((((.(...((((......))))).)))))).)))...))))))))))).))))).))...))))))..))).)).......... ( -57.00, z-score = -3.71, R) >droSec1.super_0 7240595 143 + 21120651 UCUUAGGCACUCAGUUGAG-GUUGUAGUCAUUUUG-GAACAGCAAAGUGGGGCCAGCGACUGCCCGUCACGCGACUAACUGCGCCGGCUGUGGCUUUUGCCCCACUCGCUUGUCCGAAUCAGACCUUUUACUAAGACAUCAUAGU ((((((((.(((....)))-))....(((..((((-((..(((..((((((((.(((.((.(((.(...((((......))))).))).)).)))...)))))))).)))..))))))...)))......))))))......... ( -51.20, z-score = -2.19, R) >droYak2.chr3L 15220672 143 + 24197627 UCUUGGGCACUCAGUUGAG-GUUGAAGUCAUUUUG-GGACAGCGAAGAGGAGCCAGCUACUGCCCGUCACGCGACUAACUGCGCCGGCAGUGGCUUUUGCCCAACCCGCUUGUCCGAAUCAGACCUUUUACUAAGACAUCAUAGU ((((((((.(((....)))-))....(((...((.-((((((((....((.((.((((((((((.(...((((......))))).))))))))))...))))....))).))))).))...)))......))))))......... ( -48.90, z-score = -1.56, R) >droEre2.scaffold_4784 15084255 143 + 25762168 GCUCGGGCACUCAGUUGAG-GUUGAAGUCAUUUUG-GGACAGCGAAGAGGAGCCAGCGACUGCCCGUCACGCGACUAACUGCGCCGGCGGUGGCUUUUGCCCCACCCGCUUGUCCGAAUCAGACCUUUUACUAAAACAUCAUAGU ((....)).....(((..(-((.((((((...((.-((((((((....((.((.(((.((((((.(...((((......))))).)))))).)))...))))....))).))))).))...)).)))).)))..)))........ ( -44.40, z-score = 0.25, R) >droAna3.scaffold_13337 5434314 144 - 23293914 UUUGAGGCACUCAGCAAGG-UUUGUAGUCAUUUUGAGGACAGCAAAGUGGAGCCAGCCAACGCCUGUCACGCGACUAACUGCGCCGGCGACGGCCUCCGCCCCACCUGCUCAUCCGAAUCAGACCUUUUACUAAGACAUCAUAGU .(((((...))))).((((-((((...((....((((..(((....((((((...(((..((((.(...((((......))))).))))..))))))))).....)))))))...))..))))))))..((((.(....).)))) ( -40.80, z-score = -0.32, R) >dp4.chrXR_group6 12706148 143 - 13314419 UUUUCGUUACUCAGCUGAG-AUUGUAGUCACUUUG-GGAUAGCCGAGUGGAGCCAGCCUCCGCCUGUCACGCGACUAACUGCGUCGGCGGAGCCUUUUGCUCCAUCUGCUUGUCCGAAUCAGACCUUUUACUGGGACAUCAUUGU .........(((((.((((-(.....(((...((.-(((((((.((.((((((.((.(((((((....(((((......))))).))))))).))...)))))))).)).))))).))...))).)))))))))).......... ( -49.90, z-score = -2.06, R) >droPer1.super_12 1124162 143 - 2414086 UUUUCGUUACUCAGCUGAG-AUUGUAGUCACUUUG-GGACAGCCGAGUGGAGCCAGCCUCCGCCUGUCACGCGACUAACUGCGUCGGCGGAGCCUUUUGCUCCAUCUGCUUGUCCGAAUCAGACCUUUUACUGGGACAUCAUUGU .........(((((.((((-(.....(((...((.-(((((((.((.((((((.((.(((((((....(((((......))))).))))))).))...)))))))).)).))))).))...))).)))))))))).......... ( -51.80, z-score = -2.44, R) >droMoj3.scaffold_6680 7628894 142 + 24764193 ---AUUUUACUCAGCUAAAUGUUGAAGUCAUUUUAUGAGCAGCAGAGACACGUUCGCCUGCACACAAACCGCGACUAACUGCGUCGGUGCAGGACAAGACGUGAACCGUCUGCCCAACUCAGACCUUUUACUAAAACAUCAUUGU ---.......(((((.....)))))(((.......((((..((((((.((((((..(((((((......((((......))))...)))))))....))))))...).)))))....))))........)))............. ( -36.96, z-score = -1.48, R) >droVir3.scaffold_13049 19929322 142 + 25233164 --GCUUGUACUCAGAUAAAUGUUGAAGUCAUUUUAUGGGCAGCAGAGGCACGCCCGCCUUCACCUGGCACGCGACUAACUGCGU-GGGGAAGGGCUACGUGCACACCGCAUGCCCAACUCAGACCUUUUACUAAAACAUCAUGGU --........((((((....((.((((((......((((((((.(.((((((...(((((..((...((((((......)))))-)))..)))))..))))).).).)).)))))).....)).)))).))......))).))). ( -42.70, z-score = -0.41, R) >droGri2.scaffold_15110 7119449 142 - 24565398 ---UUUUUGCUCAGCUAAAUUCUUUAGUCAUUUUGUGGCCAGCAGAGACUCAUUCGCCUUUACCUGACACGCGACUAACUGCGUGGGUGAAGAUCCCAGUGUACUCCGCUUGACCACAUCAGACCUUUUACUCAAACAUCAUAGU ---.......................(((....((((((.(((.(((...((((.(.(((((((...((((((......)))))))))))))...).))))..))).))).).)))))...)))..................... ( -33.60, z-score = -1.09, R) >consensus UCUUAGGCACUCAGCUGAG_GUUGUAGUCAUUUUG_GGACAGCAAAGUGGAGCCAGCCACUGCCCGUCACGCGACUAACUGCGCCGGCGGAGGCUUUUGCCCCACCCGCUUGUCCGAAUCAGACCUUUUACUAAGACAUCAUAGU ..........................(((.......(((((((.....(((((.....((((((.....((((......))))..)))))).......)))))....)).)))))......)))..................... (-22.57 = -21.60 + -0.97)
| Location | 15,127,642 – 15,127,785 |
|---|---|
| Length | 143 |
| Sequences | 11 |
| Columns | 145 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Shannon entropy | 0.51251 |
| G+C content | 0.51805 |
| Mean single sequence MFE | -50.40 |
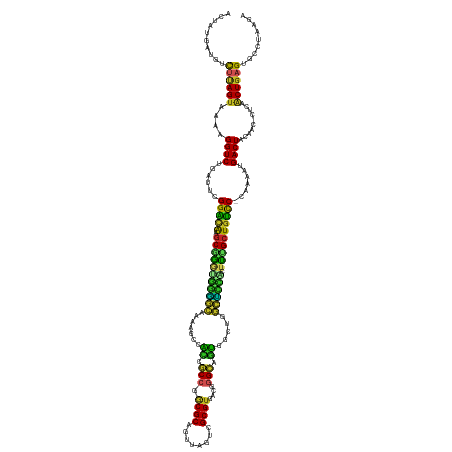
| Consensus MFE | -32.15 |
| Energy contribution | -28.94 |
| Covariance contribution | -3.21 |
| Combinations/Pair | 1.86 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
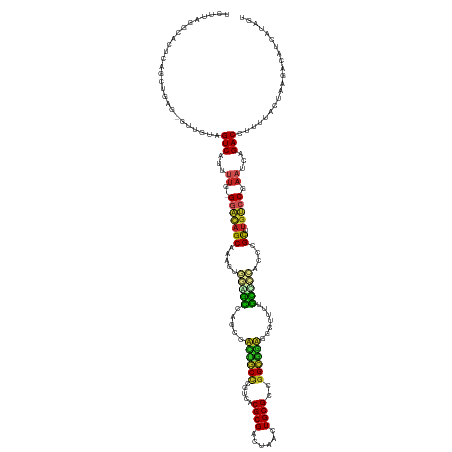
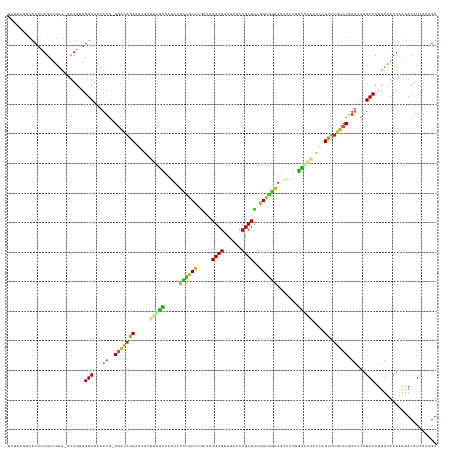
>dm3.chr3L 15127642 143 - 24543557 ACUAUGAUGUCUUAGUAAAAGGUCUGAUUCGGACAAGCGAGUGGGGCAAAAGCCACCGCCGGCGCAGUUAGUCGCGUGACGAGCAGUCGCUGGCUUCACUUCGCUGUCC-CAAAAUGACUACAAC-CUCAACUGAGUGCCUAAGA .........((((((.....((((......(((((.(((((((((((....)))...((((((((.(((.(((....))).))).).))))))).)))).)))))))))-......)))).....-(((....)))...)))))) ( -50.50, z-score = -1.88, R) >droSim1.chr3L 14459429 143 - 22553184 ACUAUGAUGUCUUAGUAAAAGGUCUGAUUCGGACAAGCGAGUGGGGCAAAAGCCACCGCCGGCGCAGUUAGUCGCGUGACGGGCAGUCGCUGGCCCCACUUCGCUGUCC-CAAAAUGACUACAAC-CUCAACUGAGUGAAUAAGA .........(((((......((((......(((((.(((((((((((...(((.((.(((.((((.((.....))))).).))).)).))).))))))).)))))))))-......))))....(-(((....))).)..))))) ( -54.00, z-score = -2.81, R) >droSec1.super_0 7240595 143 - 21120651 ACUAUGAUGUCUUAGUAAAAGGUCUGAUUCGGACAAGCGAGUGGGGCAAAAGCCACAGCCGGCGCAGUUAGUCGCGUGACGGGCAGUCGCUGGCCCCACUUUGCUGUUC-CAAAAUGACUACAAC-CUCAACUGAGUGCCUAAGA .........((((((.....((((......(((((.(((((((((((...(((.((.(((.((((.((.....))))).).))).)).))).))))))).)))))))))-......)))).....-(((....)))...)))))) ( -50.50, z-score = -1.58, R) >droYak2.chr3L 15220672 143 - 24197627 ACUAUGAUGUCUUAGUAAAAGGUCUGAUUCGGACAAGCGGGUUGGGCAAAAGCCACUGCCGGCGCAGUUAGUCGCGUGACGGGCAGUAGCUGGCUCCUCUUCGCUGUCC-CAAAAUGACUUCAAC-CUCAACUGAGUGCCCAAGA .((.((....((((((...((((.(((.(((((((.(((((..((((...(((.((((((.((((.((.....))))).).)))))).))).)).))..))))))))))-......))..)))))-))..))))))....)))). ( -48.90, z-score = -0.93, R) >droEre2.scaffold_4784 15084255 143 - 25762168 ACUAUGAUGUUUUAGUAAAAGGUCUGAUUCGGACAAGCGGGUGGGGCAAAAGCCACCGCCGGCGCAGUUAGUCGCGUGACGGGCAGUCGCUGGCUCCUCUUCGCUGUCC-CAAAAUGACUUCAAC-CUCAACUGAGUGCCCGAGC .....(((.(((.....))).)))...(((((....((.((((((((....))).))))).))(((.(((((.(((.((.((((........)))).))..))).(((.-......)))......-....))))).)))))))). ( -46.80, z-score = 0.22, R) >droAna3.scaffold_13337 5434314 144 + 23293914 ACUAUGAUGUCUUAGUAAAAGGUCUGAUUCGGAUGAGCAGGUGGGGCGGAGGCCGUCGCCGGCGCAGUUAGUCGCGUGACAGGCGUUGGCUGGCUCCACUUUGCUGUCCUCAAAAUGACUACAAA-CCUUGCUGAGUGCCUCAAA ....(((.(((((((((..((((.((....((((.((((((((((((...(((((.((((.((((........))))....)))).))))).))))))).)))))))))((.....))...)).)-)))))))))).)).))).. ( -58.20, z-score = -2.44, R) >dp4.chrXR_group6 12706148 143 + 13314419 ACAAUGAUGUCCCAGUAAAAGGUCUGAUUCGGACAAGCAGAUGGAGCAAAAGGCUCCGCCGACGCAGUUAGUCGCGUGACAGGCGGAGGCUGGCUCCACUCGGCUAUCC-CAAAGUGACUACAAU-CUCAGCUGAGUAACGAAAA .......((((((((........)))....))))).......(((((...((.(((((((.((((........))))....))))))).)).)))))((((((((....-...............-...))))))))........ ( -50.05, z-score = -2.12, R) >droPer1.super_12 1124162 143 + 2414086 ACAAUGAUGUCCCAGUAAAAGGUCUGAUUCGGACAAGCAGAUGGAGCAAAAGGCUCCGCCGACGCAGUUAGUCGCGUGACAGGCGGAGGCUGGCUCCACUCGGCUGUCC-CAAAGUGACUACAAU-CUCAGCUGAGUAACGAAAA .......((((((((........)))....))))).......(((((...((.(((((((.((((........))))....))))))).)).)))))(((((((((...-...............-..)))))))))........ ( -53.45, z-score = -2.64, R) >droMoj3.scaffold_6680 7628894 142 - 24764193 ACAAUGAUGUUUUAGUAAAAGGUCUGAGUUGGGCAGACGGUUCACGUCUUGUCCUGCACCGACGCAGUUAGUCGCGGUUUGUGUGCAGGCGAACGUGUCUCUGCUGCUCAUAAAAUGACUUCAACAUUUAGCUGAGUAAAAU--- ..........((((((.(((.((..(((((((((((.(((..(((((.(((.(((((((((((((........)))..))).)))))))))))))))...)))))))))).......))))..)).))).))))))......--- ( -46.61, z-score = -1.93, R) >droVir3.scaffold_13049 19929322 142 - 25233164 ACCAUGAUGUUUUAGUAAAAGGUCUGAGUUGGGCAUGCGGUGUGCACGUAGCCCUUCCCC-ACGCAGUUAGUCGCGUGCCAGGUGAAGGCGGGCGUGCCUCUGCUGCCCAUAAAAUGACUUCAACAUUUAUCUGAGUACAAGC-- .......(((((((((((((((((.....((((((.((((.(.((((((.(((.((((((-((((........)))))...)).))))))..))))))).))))))))))......))))).....)))).))))).)))...-- ( -53.30, z-score = -2.44, R) >droGri2.scaffold_15110 7119449 142 + 24565398 ACUAUGAUGUUUGAGUAAAAGGUCUGAUGUGGUCAAGCGGAGUACACUGGGAUCUUCACCCACGCAGUUAGUCGCGUGUCAGGUAAAGGCGAAUGAGUCUCUGCUGGCCACAAAAUGACUAAAGAAUUUAGCUGAGCAAAAA--- .......(((((.(((.(((((((...((((((((.((((((.((.(.....((((.((((((((........)))))...))).)))).....).))))))))))))))))....))))......))).))))))))....--- ( -42.10, z-score = -1.33, R) >consensus ACUAUGAUGUCUUAGUAAAAGGUCUGAUUCGGACAAGCGGGUGGGGCAAAAGCCACCGCCGGCGCAGUUAGUCGCGUGACGGGCAGUGGCUGGCUCCACUUUGCUGUCC_CAAAAUGACUACAAC_CUCAACUGAGUGCCUAAGA ..........((((((....((((......(((((.(((((((((((.......((.(((.((((........))))....))).)).....)))))).)))))))))).......))))..........))))))......... (-32.15 = -28.94 + -3.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:01 2011