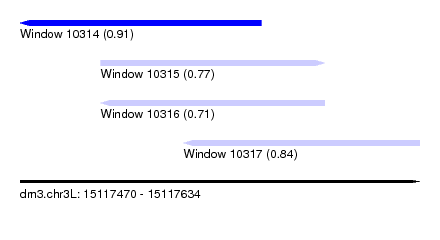
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,117,470 – 15,117,634 |
| Length | 164 |
| Max. P | 0.906863 |

| Location | 15,117,470 – 15,117,569 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Shannon entropy | 0.28576 |
| G+C content | 0.33318 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.18 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
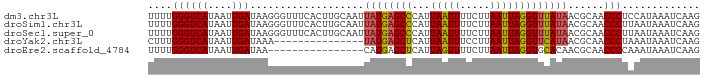
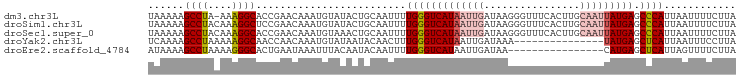
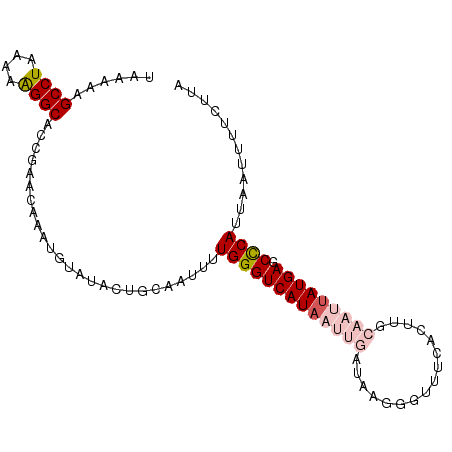
>dm3.chr3L 15117470 99 - 24543557 CACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUCCAUAAAUCAAGUUAGUAAGCUAACAAGCAAGUUGCGUGAUCUCA---- .((((((..(((((((((...(((((.....))))))))))))))....................(((((....)))))..))))))............---- ( -20.90, z-score = -2.78, R) >droSim1.chr3L 14449329 103 - 22553184 CACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUUAAUAAAUCAAGUUAGUAAGCUAACAAGCAAGUUGCGUGAUUUAAAAAA ...((((..(((((((((...(((((.....)))))))))))))).)))).......(((((((.(((((....)))))..((.....)).)))))))..... ( -22.80, z-score = -3.01, R) >droSec1.super_0 7230602 99 - 21120651 CACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUUAAUAAAUCAAGUUAGUAAGCUAACAAGCAAGUUGCGUGAUCUAA---- .((((((..(((((((((...(((((.....))))))))))))))....................(((((....)))))..))))))............---- ( -20.90, z-score = -2.46, R) >droYak2.chr3L 15210723 93 - 24197627 ----------UAUGAGCUCAUUAAUUUCCUUAAUUAGGUUCAUAACGCAACCCUAAAUAAAUCAAGUUAGUGUGUUAACAAGCCAGUUGCGAGAUCUAAUAAA ----------((((((((...(((((.....))))))))))))).((((((.((((..........))))(((....))).....))))))............ ( -16.20, z-score = -1.68, R) >droEre2.scaffold_4784 15074505 93 - 25762168 ----------CAUGAGCUCAUUAGUUUUCUUAAUUAGGUGCACAACGCAACCCCAAAUAAAUCAAGUUGGUGUGUUAACAAGCAAGUUGCGAGAUCUAAUAAA ----------.........(((((.((((.......(((((.....)).)))..........(((.(((.(((....)))..))).))).)))).)))))... ( -15.60, z-score = 0.09, R) >consensus CACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUAAAUAAAUCAAGUUAGUAAGCUAACAAGCAAGUUGCGUGAUCUAA_AAA ..........((((((((...(((((.....))))))))))))).((((((......(......)(((((....)))))......))))))............ (-14.62 = -14.18 + -0.44)
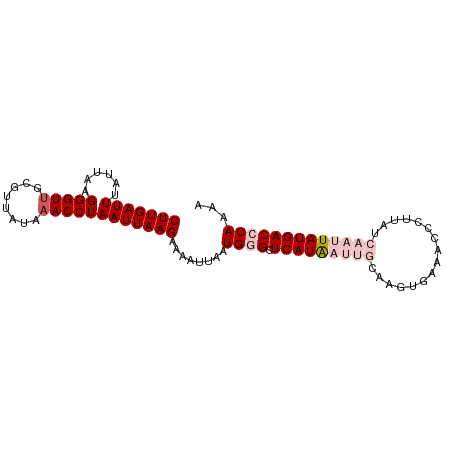
| Location | 15,117,503 – 15,117,595 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 83.54 |
| Shannon entropy | 0.27364 |
| G+C content | 0.31512 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -12.89 |
| Energy contribution | -14.93 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15117503 92 + 24543557 CUUGAUUUAUGGAGGGUUGCGUUAUAAACCUAAUUAAGAAAAUUAAUGGGCUCAUAAUUGCAAGUGAAACCCUUAUCAAUUAUGACCCAAAA (((((((......(((((........))))))))))))........((((.(((((((((.(((.(....))))..)))))))))))))... ( -22.30, z-score = -2.51, R) >droSim1.chr3L 14449366 92 + 22553184 CUUGAUUUAUUAAGGGUUGCGUUAUAAACCUAAUUAAGAAAAUUAAUGGGCUCAUAAUUGCAAGUGAAACCCUUAUCAAUUAUGACCCAAAA .(((((((.(((((((((........)))))..))))..)))))))((((.(((((((((.(((.(....))))..)))))))))))))... ( -22.40, z-score = -3.11, R) >droSec1.super_0 7230635 92 + 21120651 CUUGAUUUAUUAAGGGUUGCGUUAUAAACCUAAUUAAGAAAAUUAAUGGGCUCAUAAUUGCAAGUGAAACCCUUAUCAAUUAUGACCCAAAA .(((((((.(((((((((........)))))..))))..)))))))((((.(((((((((.(((.(....))))..)))))))))))))... ( -22.40, z-score = -3.11, R) >droYak2.chr3L 15210760 77 + 24197627 CUUGAUUUAUUUAGGGUUGCGUUAUGAACCUAAUUAAGGAAAUUAAUGAGCUCAUA---------------UUUAUCAAUUAUGACCCAAAG .(((((..((...(((((.(((((....(((.....)))....))))))))))..)---------------)..)))))............. ( -12.70, z-score = -0.88, R) >droEre2.scaffold_4784 15074542 76 + 25762168 CUUGAUUUAUUUGGGGUUGCGUUGUGCACCUAAUUAAGAAAACUAAUGAGCUCAUG----------------UUAUCAAUUAUGACCCAAAA .........(((((((((.((((((....((.....))...)).)))))))(((((----------------........))))))))))). ( -12.30, z-score = -0.10, R) >consensus CUUGAUUUAUUAAGGGUUGCGUUAUAAACCUAAUUAAGAAAAUUAAUGGGCUCAUAAUUGCAAGUGAAACCCUUAUCAAUUAUGACCCAAAA (((((((......(((((........))))))))))))........((((.(((((((((................)))))))))))))... (-12.89 = -14.93 + 2.04)

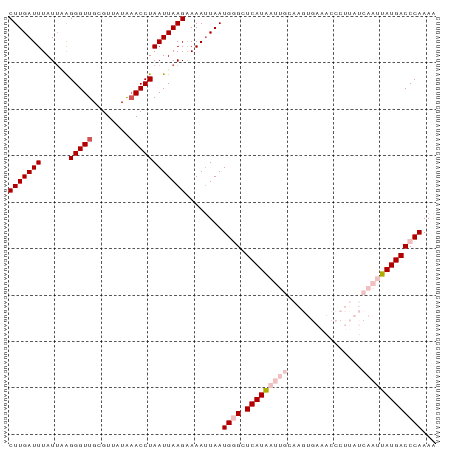
| Location | 15,117,503 – 15,117,595 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 83.54 |
| Shannon entropy | 0.27364 |
| G+C content | 0.31512 |
| Mean single sequence MFE | -17.71 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.70 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15117503 92 - 24543557 UUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUCCAUAAAUCAAG ...(((((((((((((.((((.......))))))))))))).))))................(((((((.............)))))))... ( -21.02, z-score = -2.60, R) >droSim1.chr3L 14449366 92 - 22553184 UUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUUAAUAAAUCAAG ...(((((((((((((.((((.......))))))))))))).))))................(((((((.............)))))))... ( -21.22, z-score = -2.76, R) >droSec1.super_0 7230635 92 - 21120651 UUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUUAAUAAAUCAAG ...(((((((((((((.((((.......))))))))))))).))))................(((((((.............)))))))... ( -21.22, z-score = -2.76, R) >droYak2.chr3L 15210760 77 - 24197627 CUUUGGGUCAUAAUUGAUAAA---------------UAUGAGCUCAUUAAUUUCCUUAAUUAGGUUCAUAACGCAACCCUAAAUAAAUCAAG .(((((((((....)))....---------------((((((((...(((((.....))))))))))))).......))))))......... ( -12.90, z-score = -1.93, R) >droEre2.scaffold_4784 15074542 76 - 25762168 UUUUGGGUCAUAAUUGAUAA----------------CAUGAGCUCAUUAGUUUUCUUAAUUAGGUGCACAACGCAACCCCAAAUAAAUCAAG .((((((...(((((((.((----------------((((....)))..)))...)))))))..(((.....)))..))))))......... ( -12.20, z-score = -0.91, R) >consensus UUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUAAUUAGGUUUAUAACGCAACCCUAAAUAAAUCAAG ....((((((....)))...................((((((((...(((((.....)))))))))))))......)))............. (-10.74 = -10.70 + -0.04)
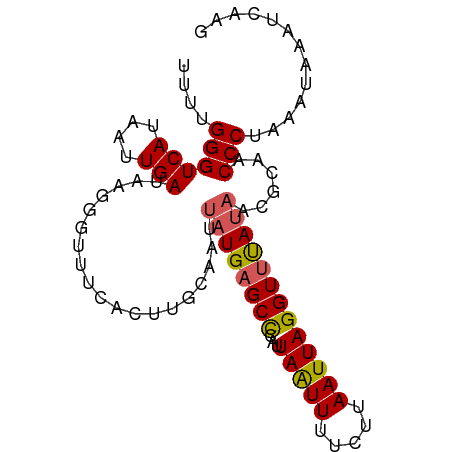
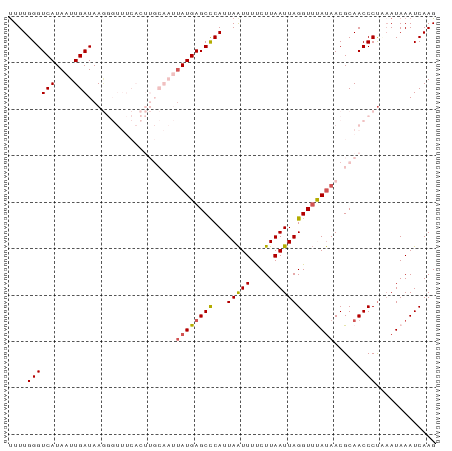
| Location | 15,117,537 – 15,117,634 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Shannon entropy | 0.32423 |
| G+C content | 0.31460 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -14.33 |
| Energy contribution | -16.48 |
| Covariance contribution | 2.15 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15117537 97 - 24543557 UAAAAAGCCUA-AAAGGCACCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUA ......(((..-...)))...(((....(((.....)))....(((((((((((((.((((.......))))))))))))).))))......)))... ( -23.70, z-score = -2.39, R) >droSim1.chr3L 14449400 98 - 22553184 UAAAAAGCCUACAAAGGCUCCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUA .....(((((....)))))..(((....(((.....)))....(((((((((((((.((((.......))))))))))))).))))......)))... ( -24.70, z-score = -2.60, R) >droSec1.super_0 7230669 98 - 21120651 UAAAAAGCCUACAAAGGCACCGAACAAAUGUAAACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUA ......((((....))))...(((....(((.....)))....(((((((((((((.((((.......))))))))))))).))))......)))... ( -24.60, z-score = -2.64, R) >droYak2.chr3L 15210794 83 - 24197627 UCAAAAGCCUAAAAAGGCAACCAACAAAUGUAUAAUACAACUUUGGGUCAUAAUUGAUAAA---------------UAUGAGCUCAUUAAUUUCCUUA (((...((((....))))..((((....((((...))))...))))((((....))))...---------------..)))................. ( -11.40, z-score = -0.73, R) >droEre2.scaffold_4784 15074576 82 - 25762168 AUAAAAGCCUAAAAGGGCACUGAAUAAAUUUACAAUACAAUUUUGGGUCAUAAUUGAUAA----------------CAUGAGCUCAUUAGUUUUCUUA ......((((....))))(((((.........(((.......))).((((....))))..----------------..........)))))....... ( -10.10, z-score = 0.22, R) >consensus UAAAAAGCCUAAAAAGGCACCGAACAAAUGUAUACUGCAAUUUUGGGUCAUAAUUGAUAAGGGUUUCACUUGCAAUUAUGAGCCCAUUAAUUUUCUUA ......((((....)))).........................(((((((((((((...(((......))).))))))))).))))............ (-14.33 = -16.48 + 2.15)
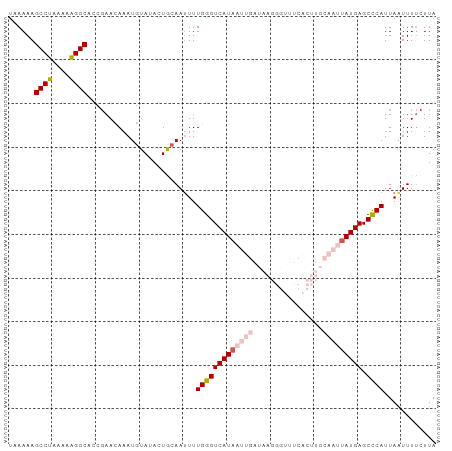
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:57 2011