| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,114,153 – 15,114,271 |
| Length | 118 |
| Max. P | 0.615657 |

| Location | 15,114,153 – 15,114,271 |
|---|---|
| Length | 118 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Shannon entropy | 0.54546 |
| G+C content | 0.57956 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -18.85 |
| Energy contribution | -18.29 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.81 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
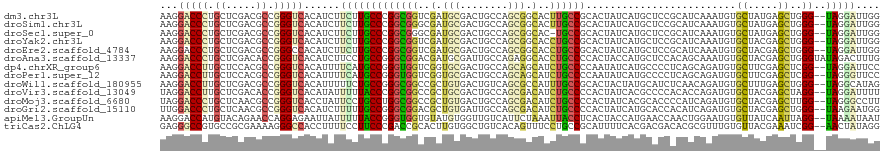
>dm3.chr3L 15114153 118 - 24543557 AAGGACCCUGCUCGACGCCGGGUCACAUCUUCUUGCCCGGCGGUCGAUGCGACUGCCAGCGGCACUUGCCGCACUAUCAUGCUCCGCAUCAAAUGUGCUAUGAGCUGGG--UAGGAUUGG ...(((((.((.....)).))))).((...((((((((((((((((...)))))))).(((((....)))))........((((.((((.....))))...)))).)))--))))).)). ( -56.60, z-score = -4.20, R) >droSim1.chr3L 14446069 118 - 22553184 AAGGACCCUGCUCGACGCCGGGUCACAUCUUCUUGCCCGGCGGGCGAUGCGACUGCCAGCGGCACUUGCCGCACUAUCAUGCUCCGCAUCAAAUGUGCUAUGAGCUGGG--UAGGAUUGG ...(((((.((.....)).))))).((...(((((((((((.((((.......)))).(((((....)))))....(((((...(((((...))))).)))))))))))--))))).)). ( -51.60, z-score = -2.40, R) >droSec1.super_0 7227354 117 - 21120651 AAGGACCCUGCUCGACGCCGGGUCACAUCUUCUUGCCCGGCGGGCGAUGCGACUGCCAGCGGCAC-UGCCGCACUAUCAUGCUCCGCAUCAAAUGUGCUAUGAGCUGGG--UAGGAUUGG ...(((((.((.....)).))))).((...(((((((((((.((((.......)))).(((((..-.)))))....(((((...(((((...))))).)))))))))))--))))).)). ( -49.70, z-score = -1.83, R) >droYak2.chr3L 15207368 118 - 24197627 AAGGACCCUGCUCGACGCCGGGUCACAUCUUCUUGCCCGGCGGUCGAUGCGACUGCCAGCGGCACCUGCCGCACUAUCAUGCUCCGCAUCAAAUGUGCUACGAGCUGGG--UAGGAUUGG ...(((((.((.....)).))))).((...((((((((((((((((...)))))))).(((((....)))))........((((.((((.....))))...)))).)))--))))).)). ( -56.60, z-score = -4.03, R) >droEre2.scaffold_4784 15071149 118 - 25762168 AAGGACCCUGCUCGACGCCGGGCCACAUCUUCUUGCCCGGCGGUCGAUGCGACUGCCAGCGGCACCUGCCGCACUAUCAUGCUCCGCAUCAAAUGUGCUACGAGCUGGG--UAGGAUUGG ..((.(((.((.....)).))))).((...((((((((((((((((...)))))))).(((((....)))))........((((.((((.....))))...)))).)))--))))).)). ( -55.10, z-score = -3.50, R) >droAna3.scaffold_13337 5419903 120 + 23293914 AAGGACCCUGCUCGACACCGGGUCACAUCUUCCUGCCGGGCGGACGAUGCGAUUGCCAGAGGCACCUGCCCCACUACCAUGCUCCACAGCAAAUGUGCUACGAGCUGGGUAUAGACUUUG ....((((.(((((.(((.(((((.((((.(((.((...))))).)))).)).((((...)))).....))).......((((....))))...)))...))))).)))).......... ( -37.60, z-score = -0.17, R) >dp4.chrXR_group6 12692883 118 + 13314419 AAGGACCUUGCUCCACGCCGGGUCACAUUUUCAUGCCGGGUGGUCGGUGCGACUGCCAGCAGCAUCUGCCCCAAUAUCAUGCCCCUCAGCAGAUGUGCUUCGAGCUCGG--UAGGAUUCC ...(((((.((.....)).)))))......((.(((((((..((((...))))..))((((.(((((((...................))))))))))).......)))--)).)).... ( -41.01, z-score = -0.23, R) >droPer1.super_12 1111329 118 + 2414086 AAGGACCUUGCUCCACGCCGGGUCACAUUUUCAUGCCGGGUGGUCGGUGCGACUGCCAGCAGCAUCUGCCCCAAUAUCAUGCCCCUCAGCAGAUGUGCUUCGAGCUCGG--UAGGGUUCC ..(((((((((.((.((((.((.((........)))).))))...)).))))(((((((((.(((((((...................)))))))))))..(....)))--)))))))). ( -41.71, z-score = -0.12, R) >droWil1.scaffold_180955 2002978 118 + 2875958 AAGGACCUUGCUCGACGCCGGGUCACAUUUUUCUGCCGGGCGGCCGCUGUGACUGUCAGCGCCAUUUGCCGCACUACUAUGCAUCUCAACAGAUGUGCUUUGAGCUGGG--UAGGCAUAG ...(((((.((.....)).))))).......((((((.((((((.((((.......))))))).................((((((....)))))).......))).))--))))..... ( -39.80, z-score = -0.02, R) >droVir3.scaffold_13049 17717165 118 - 25233164 UAGGACCUUGCUCGACACCGGGUCACAUAUUUUUACCCGGCGGCCGCUGCGACUGCCAGCGACAUCUGCCCCACUAUCACGCCCCACACCAGAUGUGCUACGAGCUAGG--UAGGAUUUU ....((((.(((((...((((((...........)))))).((((((((.(....))))))(((((((.....................)))))))))).))))).)))--)........ ( -38.00, z-score = -1.82, R) >droMoj3.scaffold_6680 22774152 118 - 24764193 UAGGACCCUGCUCAACGCCGGGUCACCUAUUCCUGCCUGGCGGCCGCUGUGACUGCCAGCGACAUCUGCCCCACUAUCACGCACCCCAUCAGAUGUGCUACGAGCUUGG--UAGGGCCUU .(((.((((((.((((((((((.((........)))))))))(((((((.(....))))))(((((((.....................))))))).......))))))--)))))))). ( -42.40, z-score = -1.12, R) >droGri2.scaffold_15110 10104247 118 + 24565398 UUGGACCCUGCUCAACGCCGGGUCACAUCUUUUUGCCGGGCGGACGCUGUGAUUGCCAGCGACAUCUGCCCCACUAUCAUGCACCACAUCAGAUGUGCUACGAGCUGGG--UAAGAAUGG .(((((((.((.....)).))))).))..((((((((((((((((((((.(....))))))...)))))))((((.....((((..........))))....)).))))--))))))... ( -42.60, z-score = -1.62, R) >apiMel3.GroupUn 10556728 118 - 399230636 AAGGACCAUGUACAGAACCAGGAGAAUUAUUUUUACCGGGUGGUGUAUGUGGUUGUCAUUCUAAAUUACCUCACUACCAUGAACCAACUGGAAUGUGUUAUCAAUUAGG--UAAAAUAAU ...(((((((((((..(((.((((((....)))).)).)))..)))))))))))...........(((((((((..(((.(......))))...))).........)))--)))...... ( -29.10, z-score = -0.78, R) >triCas2.ChLG4 6969881 118 - 13894384 GAGGGCCGUGCCGCGAAAAGGGCCACCUUUUCCUUCCCGACCGCACUUGUGGCUGUCACAGUUUCCUGCCGCAUUUUCACGACGACACGCGUUUGUGUUACGAAAUCGG--AACUAUAGG .((..((((((.(((((((((....)))))))......(((.((.(....))).)))..........)).)))......((..((((((....)))))).))....)))--..))..... ( -31.00, z-score = 1.01, R) >consensus AAGGACCCUGCUCGACGCCGGGUCACAUCUUCUUGCCCGGCGGUCGAUGCGACUGCCAGCGGCACCUGCCGCACUAUCAUGCUCCGCAUCAAAUGUGCUACGAGCUGGG__UAGGAUUGG ...(((((.((.....)).)))))......(((((((((((((..(.(((........))).)..)))))).........................((.....))..))..))))).... (-18.85 = -18.29 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:54 2011