| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,102,287 – 15,102,388 |
| Length | 101 |
| Max. P | 0.975573 |

| Location | 15,102,287 – 15,102,388 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.45 |
| Shannon entropy | 0.59671 |
| G+C content | 0.33106 |
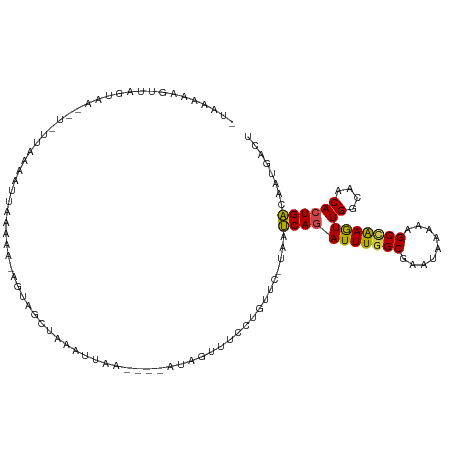
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.18 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
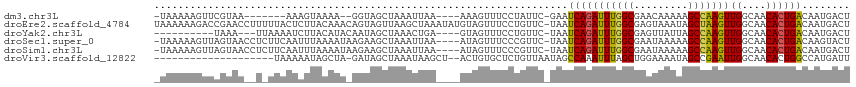
>dm3.chr3L 15102287 101 + 24543557 -UAAAAAGUUCGUAA-------AAAGUAAAA--GGUAGCUAAAUUAA----AAAGUUUCCUAUUC-GAAUCAGAUUUGGCGAACAAAAAGCCAAGUUGGCAACACUGACAAUGACU -.....(((.(((..-------........(--((.((((.......----..)))).)))....-...(((((((((((.........)))))))((....))))))..)))))) ( -18.50, z-score = -1.58, R) >droEre2.scaffold_4784 15058400 115 + 25762168 UAAAAAAGACCGAACCUUUUUACUCUUACAAACAGUAGUUAAGCUAAAUAUGUAGUUUCCUGUUC-UAAUCAGAUUUGGCGAGUAAAUAGCUAAGUUGGCAACACUGACAAUGACU .......(.((((..((.(((((((((((.....))))....(((((((...(((.........)-)).....)))))))))))))).)).....)))))................ ( -19.00, z-score = 0.26, R) >droYak2.chr3L 15195238 98 + 24197627 ----------UAAA---UUAAAAUCUUACAUACAAUAGCUAAACUGA----GUAGUUUCCUGUUC-UAAUCAGAUUUGGCGAGUUAUUAGCCAAGUUGGCAACACUGACAAUGACU ----------....---....................((((((((((----.(((.........)-)).)))).)))))).(((((((.(...(((.(....))))..)))))))) ( -18.80, z-score = -0.94, R) >droSec1.super_0 7215142 110 + 21120651 -UAAAAAGUUAGUAACCUCUUCAAUUUAAAAUAAGAAGCUAAAUUAA----AUAGUUUCCCGUUC-UAAUCAGAUUUGGCGAAUAAAAAGCCAAGUUGGCAACACUGACAAGUACU -......((((((............(((.(((..(((((((......----.)))))))..))).-)))...((((((((.........))))))))(....)))))))....... ( -21.90, z-score = -2.64, R) >droSim1.chr3L 14421928 110 + 22553184 -UAAAAAGUUAGUAACCUCUUCAAUUUAAAAUAAGAAGCUAAAUUAA----AUAGUUUCCCGUUC-UAAUCAGAUUUGGCGAAUAAAAAGCCAAGUUGGCAACACUGACAAUGACU -......((((((............(((.(((..(((((((......----.)))))))..))).-)))...((((((((.........))))))))(....)))))))....... ( -21.90, z-score = -2.59, R) >droVir3.scaffold_12822 2468896 93 - 4096053 --------------------UAAAAAUAGCUA-GAUAGCUAAAUAAGCU--ACUGUGCUCUGUUAAUAGCCAAAUUUAGCUGGAAAAUAGCCGAAUUGGCAACACUGGCCAUGAUU --------------------........((((-(.(((((.....))))--)................(((((.((..((((.....))))..)))))))....)))))....... ( -23.20, z-score = -1.61, R) >consensus _UAAAAAGUUAGUAA__U_UUAAAAUUAAAAA_AGUAGCUAAAUUAA____AUAGUUUCCUGUUC_UAAUCAGAUUUGGCGAAUAAAAAGCCAAGUUGGCAACACUGACAAUGACU .....................................................................(((((((((((.........)))))))((....))))))........ (-11.51 = -11.18 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:53 2011