| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,083,264 – 15,083,369 |
| Length | 105 |
| Max. P | 0.998611 |

| Location | 15,083,264 – 15,083,369 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 55.75 |
| Shannon entropy | 0.94737 |
| G+C content | 0.40568 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -9.95 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
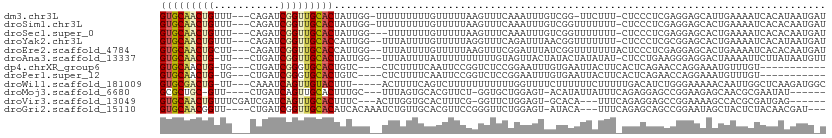
>dm3.chr3L 15083264 105 - 24543557 GUGCAACUGUUU---CAGAUCGGUUGCACUAUUGG-UUUUUUUUUGUUUUUAAGUUUCAAAUUUGUCGG-UUCUUU-CUCCCUCGAGGAGCAUUGAAAAUCACAUAAUGAU (((((((((...---.....)))))))))...(((-(((((...(((((............((((..((-......-..))..)))))))))..))))))))......... ( -23.40, z-score = -1.56, R) >droSim1.chr3L 14402789 106 - 22553184 GUGCAACUGUUU---CAGAUCGGUUGCACUAUUGG-UUUUUUUUUGUUUUUAAGUUUCAAAUUUGUCGGUUUUUUU-CUCCCUCGAGGAGCACUGAAAAUCACACAAUGAU (((((((((...---.....)))))))))(((((.-......((((...........))))..(((.(((((((..-((((.....))))....))))))))))))))).. ( -25.20, z-score = -1.86, R) >droSec1.super_0 7197387 104 - 21120651 GUGCAACUGUUU---CAGAUCGGUUGCACUAUUGG---UUUUUUUGUUUUUAAGUUUCAAAUUUGUCGGUUUUUUU-CUCCCUCGAGGAGCACUGAAAAUCACACAAUGAU (((((((((...---.....)))))))))(((((.---....((((...........))))..(((.(((((((..-((((.....))))....))))))))))))))).. ( -25.20, z-score = -1.83, R) >droYak2.chr3L 15176634 105 - 24197627 GUGCAACUGUUU---CAGAUCGGUUGCACCAUUGG--UUUAUUUUGUUUUUAGGUUUCAGAUUUAACGGUUUUUUU-CUCCCUCGCGGAGCACUGAAAAUCACAUAAUGAU (((((((((...---.....)))))))))((((((--((...((((...........))))...)))(((((((..-((((.....))))....)))))))...))))).. ( -26.30, z-score = -2.14, R) >droEre2.scaffold_4784 15039816 106 - 25762168 GUGCAACUGCUU---CAGAUCGGUUGCACCAUUGG--UUUAUUUUGUUUUUAAGUUUCGGAUUUAUCGGUUUUUUUACUCCCUCGAGGAGCACUGAAAAUCACACAAUGAU (((((((((...---.....)))))))))(((((.--.............((((((...))))))..(((((((...((((.....))))....)))))))...))))).. ( -25.90, z-score = -1.82, R) >droAna3.scaffold_13337 5391259 104 + 23293914 GUGCAACUG-UU---CUGAUCGGUUGCACUAUUGG--UUUAUUUUAUUUUUUUUUUGUAGUUACUAUACUAUAUAU-CUCCUGAAGGGAGGACUAAAAUUCUUAUAAUGUU (((((((((-..---.....)))))))))..((((--((................((((((......))))))...-((((.....))))))))))............... ( -25.80, z-score = -3.66, R) >dp4.chrXR_group6 12659008 92 + 13314419 GUGCAACUG-UG---CUGAUCGGGUGCACUGUC----CUCUUUUCAAUUCCGGUCUCCGGAAUUUGUGAAUUACUUCACUCAGAACCAGGAAAUGUUUGU----------- (((((.(((-..---.....))).)))))..((----((...(((((((((((...)))))))).(((((....)))))...)))..)))).........----------- ( -26.20, z-score = -1.77, R) >droPer1.super_12 1076820 92 + 2414086 GUGCAACUG-UG---CUGAUCGGGUGCACUGUC----CUCUUUUCAAUUCCGGUCUCCGGAAUUUGUGAAUUACUUCACUCAGAACCAGGAAAUGUUUGU----------- (((((.(((-..---.....))).)))))..((----((...(((((((((((...)))))))).(((((....)))))...)))..)))).........----------- ( -26.20, z-score = -1.77, R) >droWil1.scaffold_181009 649291 102 + 3585778 GUGCGACUG-UU---CAAAUCAGUUGUACUUU-----ACUUUUCAGUCUUUUUUUUUUUGGUUUUCUUUUUUCUUUUUGACAUCUGGGAAAAACAAUUGGCUCAAGAUGGC (((((((((-..---.....)))))))))...-----..((((.((((...(((((((..(...((............))...)..))))))).....)))).)))).... ( -20.30, z-score = -1.36, R) >droMoj3.scaffold_6680 22739136 95 - 24764193 GCGCUGC-GUU----CUGAUCAGUUGCACUUUGC---UUUAGUGCACGUUCU-GGUGCUGGAGU-ACAUAUUAUUUCAGAGGAGCCGGAAGAGCAACGCGAAUAU------ (((.(((-.((----(((....((.(((((....---...)))))))(((((-....(((((((-(.....)))))))).))))))))))..))).)))......------ ( -32.80, z-score = -1.82, R) >droVir3.scaffold_13049 17688253 97 - 25233164 GUGCAACUGUUUCGAUCGAUCAGUUGCACUUUC---ACUUGGUGCACUUUCG-GGUUCUGGAGU-GCACA---UUUCAGAGGAGCCGGAAAAGCCACGCGAUGAG------ (((((((((..((....)).)))))))))..((---.(.((((....((((.-(((((((((((-....)---)))))))...))).)))).)))).).))....------ ( -35.60, z-score = -2.40, R) >droGri2.scaffold_15110 10072741 100 + 24565398 GUGCAACGGUU----CUGAUCGGUUGCACAUCACAAAUCUGUUGCACGUUCCGGGUUCUGGAGU-AUACA---UUUCAGAGCAGCCGGAAUAGCUACUCUACAACGAU--- (((((((((..----.((((.(......))))).....)))))))))(((((((((((((((((-....)---)))))))))..))))))).................--- ( -36.30, z-score = -3.93, R) >consensus GUGCAACUGUUU___CAGAUCGGUUGCACUAUUGG__UUUUUUUUAUUUUUAAGUUUCAGAUUUUACGGUUUAUUU_CUCCCUCGCGGAGCACUGAAAAUCACACAAUG_U (((((((((...........))))))))).................................................................................. ( -9.95 = -10.13 + 0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:48 2011