| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,055,437 – 15,055,544 |
| Length | 107 |
| Max. P | 0.842907 |

| Location | 15,055,437 – 15,055,544 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.61 |
| Shannon entropy | 0.68247 |
| G+C content | 0.37545 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -6.62 |
| Energy contribution | -6.33 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.64 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15055437 107 - 24543557 AUGUGUGCACGAAAAGAAAGUCAAUUUAAUUGUUUAAAC--UCUCAGUUGGUCGUCUACGUAUUUGCAAAAACU--UUCAA-----ACUGUCACCUGCAGUCGUGCAUACAAAGUU---- .(((((((((((...((((((...(((((....))))).--.....((..((((....)).))..))....)))--)))..-----.((((.....))))))))))))))).....---- ( -27.70, z-score = -2.91, R) >droEre2.scaffold_4784 15013091 90 - 25762168 ------GCUCGCAGAAGAAGACAGUUUAUUUGCUUGAAU--UCGCAGUUGGUCCUCUUCGCAUUUGCAAUAG-U--UUCCA-----ACUGCCACCUACAAAAAAUU-------------- ------....(((((.(((.....))).)))))......--..((((((((....((..((....))...))-.--..)))-----)))))...............-------------- ( -19.30, z-score = -1.11, R) >droYak2.chr3L 15149400 106 - 24197627 AUGUGUGCACGUAGGAGAAGACAAUUUAAUUGCUCAAAU--UCUCACCCCUUCUUUU-CGUAUUUGCAAAAACU--UUCCA-----ACUGUCAGCUGCAGUCGUGUAUACAAAGUU---- .((((((((((..(((((.((.(((((........))))--).))............-.((....))......)--)))).-----(((((.....))))))))))))))).....---- ( -22.60, z-score = -1.48, R) >droSec1.super_0 7170893 107 - 21120651 AUGUGUGCACGAAAAAGAAGCCAACUUAAUUGUUUAAAC--UCUCAGUUGGUCUUCUACGUAUUUGCAAAAACU--UUCAA-----ACUGUCACCUGCACUCGUGCAUACAAAGUU---- .(((((((((((...(((((((((((.....((....))--....)))))).))))).......((((...((.--.....-----...))....)))).))))))))))).....---- ( -31.40, z-score = -4.51, R) >droSim1.chr3L 14376366 107 - 22553184 AUGUGUGCACGAAAAAGAAGUCAAUUUAAUUGUUUAAAC--UCUCAGUUGGUCUUCUACGUAUUUGCAAAAACU--UUCAA-----ACUGUCACCUGCACUCGUGCAUACAAAGUU---- .(((((((((((...(((((((((((.....((....))--....)))))).))))).......((((...((.--.....-----...))....)))).))))))))))).....---- ( -26.40, z-score = -2.89, R) >dp4.chrXR_group6 12631955 119 + 13314419 -CCGUUGGAUACAUCAGAAAUGGAUUACUACGGCUAAGCGACACUACUAUGCUGUGGAUAUAUUAGAGAAAAUUAAUUACAAAUGUAUUGCCAUCUAUGUAGAUCUUAGCGAAUGGAGAU -(((((.(.(((((.(((..(((....(((((((..((........))..)))))))(((((((.(.((.......)).).)))))))..)))))))))))........).))))).... ( -21.70, z-score = 0.01, R) >consensus AUGUGUGCACGAAAAAGAAGUCAAUUUAAUUGUUUAAAC__UCUCAGUUGGUCUUCUACGUAUUUGCAAAAACU__UUCAA_____ACUGUCACCUGCACUCGUGCAUACAAAGUU____ .((((((((((................................................((....)).....................((.......))..))))))))))......... ( -6.62 = -6.33 + -0.29)

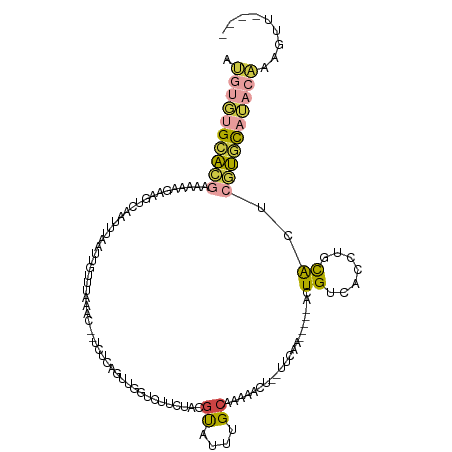

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:44 2011