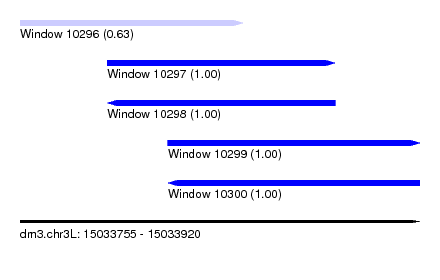
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,033,755 – 15,033,920 |
| Length | 165 |
| Max. P | 0.999555 |

| Location | 15,033,755 – 15,033,847 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.61 |
| Shannon entropy | 0.62398 |
| G+C content | 0.52834 |
| Mean single sequence MFE | -29.09 |
| Consensus MFE | -11.87 |
| Energy contribution | -12.11 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
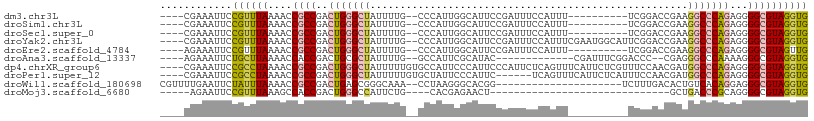
>dm3.chr3L 15033755 92 + 24543557 ----CGAAAUUCCGUUUAAAACCGCCGACUGGGCUAUUUUG--CCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGGUG ----................((((((((.(((((......)--)))))))))..(((((.........----------)))))......((((....))))...))). ( -33.20, z-score = -1.91, R) >droSim1.chr3L 14354889 92 + 22553184 ----CGAAAUUCCGUUUAAAACCGCCGACUGGGCUAUUUUG--CCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGGUG ----................((((((((.(((((......)--)))))))))..(((((.........----------)))))......((((....))))...))). ( -33.20, z-score = -1.91, R) >droSec1.super_0 7150361 92 + 21120651 ----CGAAAUUCCGUUUAAAACCGCCGACUGGGCUAUUUUG--CCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGGUG ----................((((((((.(((((......)--)))))))))..(((((.........----------)))))......((((....))))...))). ( -33.20, z-score = -1.91, R) >droYak2.chr3L 15125873 102 + 24197627 ----CGAAAUUCCGUUUAAAACCGCCGACUGGGCUAUUUUG--CCCAUUGGCAUUCCGAUUUCCAUUUCGAAUGGCAUUCGGACCGAAGGCCCAGAGGGGCGUAGGUG ----................((((((((.(((((......)--)))))))))..(((((...((((.....))))...)))))......((((....))))...))). ( -37.70, z-score = -2.10, R) >droEre2.scaffold_4784 14989652 92 + 25762168 ----AGAAAUUCCGUUUAAAACCGCCGACUGGGCUAUUUUG--CCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGUUG ----.....(((...........(((((.(((((......)--)))))))))..(((((.........----------)))))..))).((((....))))....... ( -30.70, z-score = -1.70, R) >droAna3.scaffold_13337 5342113 87 - 23293914 ----AGAAAUUCUGCUUAAAACCACCGACUGCGCUAUUUUG--GCCAUUCGCAUAC-------------CGAUUUCGGACCC--CGAGGGCCCAAAAGGGCGUAGGUG ----..................((((...((((((.(((((--(((.((((....(-------------((....)))....--)))))).)))))).)))))))))) ( -28.50, z-score = -1.89, R) >dp4.chrXR_group6 12612517 104 - 13314419 ----CGAAAUUCCGCCUAAAACCGCCGACUGGGCUAUUUUUGUGCCAUUCCCAUUCCCAUUCUCAGUUUCAUUCUCGUUUCCAACGAUGGCCCAGAGGGGCGUAGGUG ----........((((((...((.((..((((((((((.(((.......................................))).)))))))))).))))..)))))) ( -27.93, z-score = -0.85, R) >droPer1.super_12 1029625 98 - 2414086 ----CGAAAUUCCGCCUAAAACCGCCGACUGGGCUAUUUUUGUGCUAUUCCCAUUC------UCAGUUUCAUUCUCAUUUCCAACGAUGGCCCAGAGGGGCGUAGGUG ----........((((((...((.((..((((((((((.(((..............------...................))).)))))))))).))))..)))))) ( -28.11, z-score = -1.48, R) >droWil1.scaffold_180698 5114626 85 - 11422946 CGUUUUGAAUUCUAUUUAAAACCGCCGACUGAGCGGGCAAA--CCUAAGGGCACGG---------------------UCUUUGACACUGUCACAGGAGGGCGUAGGUG .(((((((((...)))))))))(((((((((.(((((....--)))....)).)))---------------------))....((.((.((....)).)).)).)))) ( -22.20, z-score = 0.02, R) >droMoj3.scaffold_6680 22696646 69 + 24764193 -----AGAAUUCCGUUUAAAGCCACCGACUGGGCCAUUCUG----CACGAGAACU------------------------------GCUGACCCGCAGGGGCGUAGGUG -----.................((((...((.(((.((((.----....))))((------------------------------((......)))).))).)))))) ( -16.20, z-score = 1.23, R) >consensus ____CGAAAUUCCGUUUAAAACCGCCGACUGGGCUAUUUUG__CCCAUUGGCAUUCCGAUUUCCAUUU__________UCGGACCGAAGGCCCAGAGGGGCGUAGGUG ............((((((....((((..(((((((.....................................................)))))))...)))))))))) (-11.87 = -12.11 + 0.24)
| Location | 15,033,791 – 15,033,885 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.54 |
| Shannon entropy | 0.64018 |
| G+C content | 0.55355 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.997030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15033791 94 + 24543557 --GCCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGCA-- --((((.((((...(((((.........----------))))))))).))((((.(((((.((.((((((((.......))).))))))).)))))))))...)).-- ( -37.80, z-score = -2.62, R) >droSim1.chr3L 14354925 94 + 22553184 --GCCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGCA-- --((((.((((...(((((.........----------))))))))).))((((.(((((.((.((((((((.......))).))))))).)))))))))...)).-- ( -37.80, z-score = -2.62, R) >droSec1.super_0 7150397 94 + 21120651 --GCCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGCA-- --((((.((((...(((((.........----------))))))))).))((((.(((((.((.((((((((.......))).))))))).)))))))))...)).-- ( -37.80, z-score = -2.62, R) >droYak2.chr3L 15125909 106 + 24197627 --GCCCAUUGGCAUUCCGAUUUCCAUUUCGAAUGGCAUUCGGACCGAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGACUCGCGCA --(((..((((...(((((...((((.....))))...))))))))).)))(((.(((((.((.((((((((.......))).))))))).))))))))......... ( -39.20, z-score = -1.68, R) >droEre2.scaffold_4784 14989688 94 + 25762168 --GCCCAUUGGCAUUCCGAUUUCCAUUU----------UCGGACCGAAGGCCCAGAGGGGCGUAGUUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGCCCGCA-- --((...((((...(((((.........----------))))))))).((((((.(((((.(..((.(((((.......))).))))..).))))))))))).)).-- ( -36.60, z-score = -2.41, R) >droAna3.scaffold_13337 5342149 89 - 23293914 --GGCCAUUCGCAUACCGAUUUCGGACC---------------CCGAGGGCCCAAAAGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUAGCA-- --........((...(((....)))..(---------------((((((((((....))))...(((((...(((........)))..))))))))))))...)).-- ( -38.30, z-score = -3.12, R) >dp4.chrXR_group6 12612553 106 - 13314419 UGUGCCAUUCCCAUUCCCAUUCUCAGUUUCAUUCUCGUUUCCAACGAUGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUGGCA-- ..(((((((((..............(..(((...((((.....)))))))..).((((((.((.((((((((.......))).))))))).)))))))))))))))-- ( -36.90, z-score = -2.40, R) >droPer1.super_12 1029661 100 - 2414086 ------UGUGCUAUUCCCAUUCUCAGUUUCAUUCUCAUUUCCAACGAUGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUGGCA-- ------..(((((((((.................(((((......)))))....((((((.((.((((((((.......))).))))))).)))))))))))))))-- ( -33.00, z-score = -1.65, R) >droWil1.scaffold_180698 5114666 82 - 11422946 ------------------ACCUAAGGGCACG-----GUCUUUGACACUGUCACAGGAGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCG-AGUACGAGGACUCGGA-- ------------------.(((...((((..-----(((...)))..)))).....)))(((((..(((....)))..)))..)).(((-(((......)))))).-- ( -24.50, z-score = -0.75, R) >droMoj3.scaffold_6680 22696681 72 + 24764193 ----------------------------------GCACGAGAACUGCUGACCCGCAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUCAUUCUUUCU-- ----------------------------------....(((..((((......))))(((.((.((((((((.......))).))))))).)))))).........-- ( -22.80, z-score = -1.49, R) >consensus __GCCCAUUGGCAUUCCGAUUUCCAUUU__________UCGGACCGAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGCA__ ...................................................(((.(((((.((.((((((((.......))).))))))).))))))))......... (-17.74 = -18.59 + 0.85)
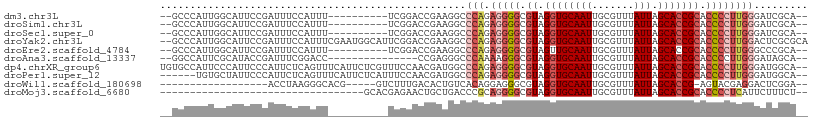
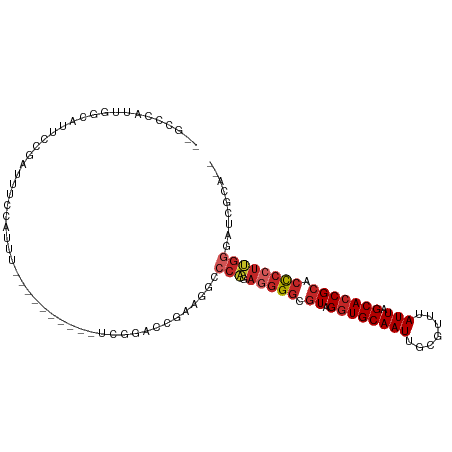
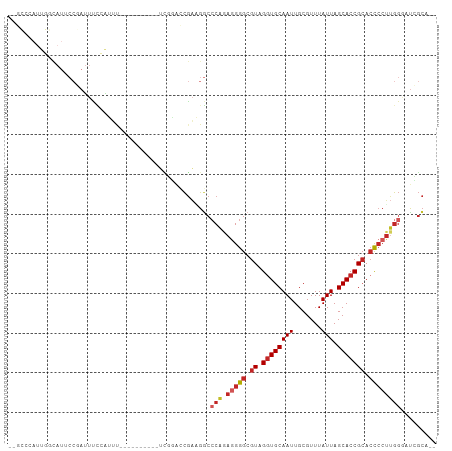
| Location | 15,033,791 – 15,033,885 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.54 |
| Shannon entropy | 0.64018 |
| G+C content | 0.55355 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.29 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15033791 94 - 24543557 --UGCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUCGGUCCGA----------AAAUGGAAAUCGGAAUGCCAAUGGGC-- --..(((.(((((((((((.(((((..............)))))..))))))).))))...))).(((((----------.........)))))..(((....)))-- ( -36.34, z-score = -1.90, R) >droSim1.chr3L 14354925 94 - 22553184 --UGCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUCGGUCCGA----------AAAUGGAAAUCGGAAUGCCAAUGGGC-- --..(((.(((((((((((.(((((..............)))))..))))))).))))...))).(((((----------.........)))))..(((....)))-- ( -36.34, z-score = -1.90, R) >droSec1.super_0 7150397 94 - 21120651 --UGCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUCGGUCCGA----------AAAUGGAAAUCGGAAUGCCAAUGGGC-- --..(((.(((((((((((.(((((..............)))))..))))))).))))...))).(((((----------.........)))))..(((....)))-- ( -36.34, z-score = -1.90, R) >droYak2.chr3L 15125909 106 - 24197627 UGCGCGAGUCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUCGGUCCGAAUGCCAUUCGAAAUGGAAAUCGGAAUGCCAAUGGGC-- ....(((((((((((((((.(((((..............)))))..))))))).)))))..))).(((((...((((.....))))...)))))..(((....)))-- ( -42.14, z-score = -2.06, R) >droEre2.scaffold_4784 14989688 94 - 25762168 --UGCGGGCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCAACUACGCCCCUCUGGGCCUUCGGUCCGA----------AAAUGGAAAUCGGAAUGCCAAUGGGC-- --.(.((((((((((((((..((((..............)).))..))))))).))))))).)..(((((----------.........)))))..(((....)))-- ( -39.14, z-score = -2.17, R) >droAna3.scaffold_13337 5342149 89 + 23293914 --UGCUAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCUUUUGGGCCCUCGG---------------GGUCCGAAAUCGGUAUGCGAAUGGCC-- --.((((((((.(((((((((((((........)).))))))))...((((....))))))).))---------------)..(((....))).......))))).-- ( -36.40, z-score = -1.85, R) >dp4.chrXR_group6 12612553 106 + 13314419 --UGCCAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCAUCGUUGGAAACGAGAAUGAAACUGAGAAUGGGAAUGGGAAUGGCACA --((((((((((..(((((((((((........)).)))))))))...(((.(((.((.(((..(((....)))..)))...)).)))..)))..)))).)))))).. ( -43.10, z-score = -3.55, R) >droPer1.super_12 1029661 100 + 2414086 --UGCCAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCAUCGUUGGAAAUGAGAAUGAAACUGAGAAUGGGAAUAGCACA------ --(((..(((((.((((((.(((((..............)))))..))))))(((.(.(((....)))...).))).............)))))...)))..------ ( -33.44, z-score = -1.79, R) >droWil1.scaffold_180698 5114666 82 + 11422946 --UCCGAGUCCUCGUACU-CGGUGCUAAUAAACGCAAUUGCACCUACGCCCUCCUGUGACAGUGUCAAAGAC-----CGUGCCCUUAGGU------------------ --.((((((......)))-)))(((........))).....((((((((..((...((((...))))..)).-----.)))....)))))------------------ ( -17.60, z-score = -0.26, R) >droMoj3.scaffold_6680 22696681 72 - 24764193 --AGAAAGAAUGAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUGCGGGUCAGCAGUUCUCGUGC---------------------------------- --.......(((((((.((((((((..............)))))...((((....))))..))).)))))))..---------------------------------- ( -24.94, z-score = -1.24, R) >consensus __UGCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUCGGUCCGA__________AAAUGGAAAUCGGAAUGCCAAUGGGC__ ........(((((((((((.(((((..............)))))..))))))).)))).................................................. (-20.84 = -21.29 + 0.45)
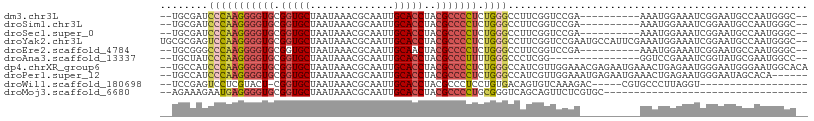
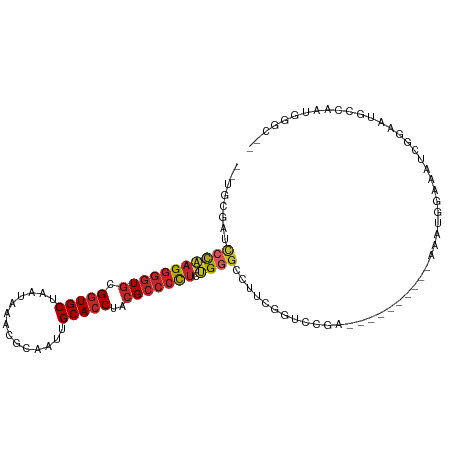
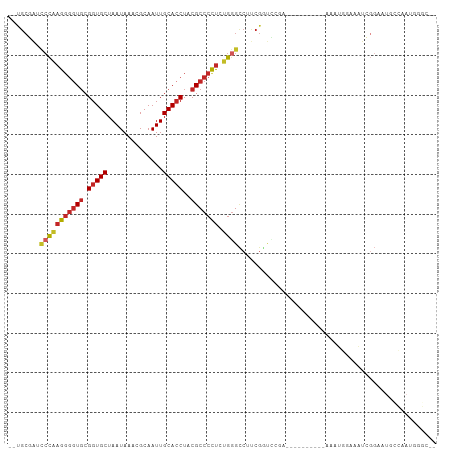
| Location | 15,033,816 – 15,033,920 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.58386 |
| G+C content | 0.54106 |
| Mean single sequence MFE | -37.96 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
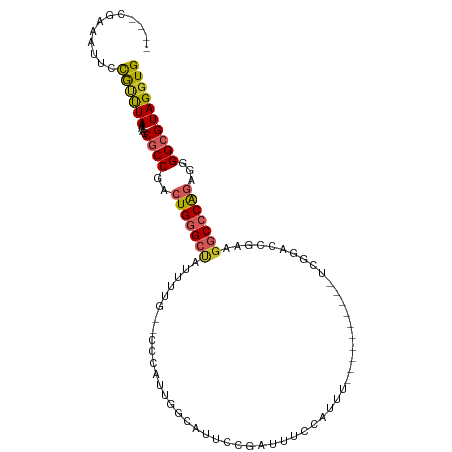
>dm3.chr3L 15033816 104 + 24543557 ----UUCGGACC--GAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGC--ACAAAAGGAUG-AGUUAGUCCUCGU-UUCGGUUCCAAG-- ----...(((((--(((((((((.(((((.((.((((((((.......))).))))))).))))))))).))..--.....(((((.-.....)))))...-))))))))....-- ( -42.10, z-score = -3.36, R) >droSim1.chr3L 14354950 104 + 22553184 ----UUCGGACC--GAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGC--ACAAAAGGAUG-AGUUAGUCCUCGU-UUCGGUUCCAAG-- ----...(((((--(((((((((.(((((.((.((((((((.......))).))))))).))))))))).))..--.....(((((.-.....)))))...-))))))))....-- ( -42.10, z-score = -3.36, R) >droSec1.super_0 7150422 104 + 21120651 ----UUCGGACC--GAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGC--ACAAAAGGAUG-AGUUAGUCCUCGU-UUCGGUUCCAAG-- ----...(((((--(((((((((.(((((.((.((((((((.......))).))))))).))))))))).))..--.....(((((.-.....)))))...-))))))))....-- ( -42.10, z-score = -3.36, R) >droYak2.chr3L 15125941 109 + 24197627 -GCAUUCGGACC--GAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGACUCGCGCACAAAAGGAUG-AGUUAGUCCUUGU-UUCGGUUCCAAG-- -......(((((--((((.....((((((.((.((((((((.......))).))))))).))))))(((((.((.((........))-.)).)))))...)-))))))))....-- ( -40.80, z-score = -2.43, R) >droEre2.scaffold_4784 14989713 104 + 25762168 ----UUCGGACC--GAAGGCCCAGAGGGGCGUAGUUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGCCCGC--ACAAAAGGAUG-AGUUAGUCCUCGU-UUCGGUUCCGAG-- ----.(((((((--(((((((((.(((((.(..((.(((((.......))).))))..).)))))))))))...--.....(((((.-.....)))))...-))))).))))).-- ( -45.70, z-score = -4.14, R) >droAna3.scaffold_13337 5342167 110 - 23293914 ----UUCGGACCCCGAGGGCCCAAAAGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUAGC--ACAAUGUCCUGCAUUCAAUGUCCGUGUUCGGUUCGGAUUC ----((((((((.(((((((((....))))...(((((...(((........)))..))))))))))(((((..--..((((.....))))...)))))......))))))))... ( -43.00, z-score = -2.67, R) >dp4.chrXR_group6 12612586 102 - 13314419 CUCGUUUCCAAC--GAUGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUGGC--ACAAUGAG----AGUUAGUCCCAGU-UCCAGGUCC----- .((((.....))--)).((((..((((((.((.((((((((.......))).))))))).))))))(((((((.--(((((...----.))).)).))).)-))).)))).----- ( -36.80, z-score = -1.99, R) >droPer1.super_12 1029688 102 - 2414086 CUCAUUUCCAAC--GAUGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUGGC--ACAAUGAG----AGUUAGUCCCAGU-UCCAGGUCC----- ............--...((((..((((((.((.((((((((.......))).))))))).))))))(((((((.--(((((...----.))).)).))).)-))).)))).----- ( -34.90, z-score = -1.60, R) >droWil1.scaffold_180698 5114681 92 - 11422946 ----CUUUGACA---CUGUCACAGGAGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCG-AGUACGAGGACUCGG-----ACCCCUGGGAUUCAAUUUUUGUAGUU----------- ----..((((..---..(((.((((.(((((((..(((....)))..)))..)).(((-(((......))))))-----.)))))).)))))))...........----------- ( -27.80, z-score = -1.45, R) >droMoj3.scaffold_6680 22696691 81 + 24764193 -------------UGCUGACCCGCAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUCAUUCUUUC--------UCUCUCAUUGAGUUUUGGAA-------------- -------------(((......)))((((.((.((((((((.......))).))))))).))))...((((...--------.(((.....)))....))))-------------- ( -24.30, z-score = -1.46, R) >consensus ____UUCGGACC__GAAGGCCCAGAGGGGCGUAGGUGCAAUUGCGUUUAUUAGCACCGCACCCCUUGGGAUCGC__ACAAAAGGAUG_AGUUAGUCCUCGU_UUCGGUUCC_AG__ ....................(((.(((((.((.((((((((.......))).))))))).))))))))................................................ (-17.74 = -18.59 + 0.85)


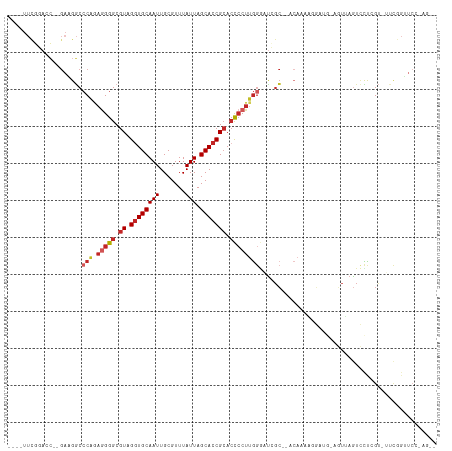
| Location | 15,033,816 – 15,033,920 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.58386 |
| G+C content | 0.54106 |
| Mean single sequence MFE | -37.79 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.29 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
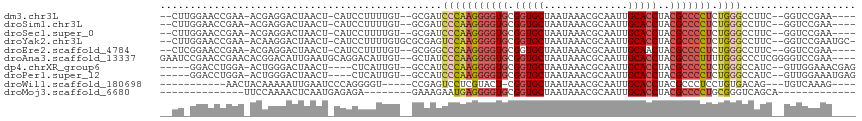
>dm3.chr3L 15033816 104 - 24543557 --CUUGGAACCGAA-ACGAGGACUAACU-CAUCCUUUUGU--GCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUC--GGUCCGAA---- --.(((((.(((((-..(((......))-)..........--.....(((((((((((.(((((..............)))))..))))))).))))..)))--))))))).---- ( -41.34, z-score = -3.76, R) >droSim1.chr3L 14354950 104 - 22553184 --CUUGGAACCGAA-ACGAGGACUAACU-CAUCCUUUUGU--GCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUC--GGUCCGAA---- --.(((((.(((((-..(((......))-)..........--.....(((((((((((.(((((..............)))))..))))))).))))..)))--))))))).---- ( -41.34, z-score = -3.76, R) >droSec1.super_0 7150422 104 - 21120651 --CUUGGAACCGAA-ACGAGGACUAACU-CAUCCUUUUGU--GCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUC--GGUCCGAA---- --.(((((.(((((-..(((......))-)..........--.....(((((((((((.(((((..............)))))..))))))).))))..)))--))))))).---- ( -41.34, z-score = -3.76, R) >droYak2.chr3L 15125941 109 - 24197627 --CUUGGAACCGAA-ACAAGGACUAACU-CAUCCUUUUGUGCGCGAGUCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUC--GGUCCGAAUGC- --.(((((.(((((-(((((((......-..))))..)))......((((((((((((.(((((..............)))))..))))))).))))).)))--)))))))....- ( -42.34, z-score = -3.34, R) >droEre2.scaffold_4784 14989713 104 - 25762168 --CUCGGAACCGAA-ACGAGGACUAACU-CAUCCUUUUGU--GCGGGCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCAACUACGCCCCUCUGGGCCUUC--GGUCCGAA---- --.(((((.(((((-..(((......))-)..........--...(((((((((((((..((((..............)).))..))))))).)))))))))--))))))).---- ( -47.84, z-score = -4.71, R) >droAna3.scaffold_13337 5342167 110 + 23293914 GAAUCCGAACCGAACACGGACAUUGAAUGCAGGACAUUGU--GCUAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCUUUUGGGCCCUCGGGGUCCGAA---- ................(((((.......(((.(....).)--))...(((.(((((((((((((........)).))))))))...((((....))))))).))))))))..---- ( -40.60, z-score = -2.12, R) >dp4.chrXR_group6 12612586 102 + 13314419 -----GGACCUGGA-ACUGGGACUAACU----CUCAUUGU--GCCAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCAUC--GUUGGAAACGAG -----((.((..((-..(((((...((.----......))--....))))).((((((.(((((..............)))))..))))))))..))))...--.(((....))). ( -38.94, z-score = -2.77, R) >droPer1.super_12 1029688 102 + 2414086 -----GGACCUGGA-ACUGGGACUAACU----CUCAUUGU--GCCAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCAUC--GUUGGAAAUGAG -----((.((..((-..(((((...((.----......))--....))))).((((((.(((((..............)))))..))))))))..)))).((--((.....)))). ( -36.44, z-score = -2.01, R) >droWil1.scaffold_180698 5114681 92 + 11422946 -----------AACUACAAAAAUUGAAUCCCAGGGGU-----CCGAGUCCUCGUACU-CGGUGCUAAUAAACGCAAUUGCACCUACGCCCUCCUGUGACAG---UGUCAAAG---- -----------...........((((.((.((((((.-----.(((((......)))-)(((((..............)))))...)..)))))).))...---..))))..---- ( -23.04, z-score = -1.89, R) >droMoj3.scaffold_6680 22696691 81 - 24764193 --------------UUCCAAAACUCAAUGAGAGA--------GAAAGAAUGAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUGCGGGUCAGCA------------- --------------.(((....(((.....))).--------.........(((((((.(((((..............)))))..)))))))..)))......------------- ( -24.64, z-score = -1.51, R) >consensus __CU_GGAACCGAA_ACGAGGACUAACU_CAUCCUUUUGU__GCGAUCCCAAGGGGUGCGGUGCUAAUAAACGCAAUUGCACCUACGCCCCUCUGGGCCUUC__GGUCCGAA____ ...............................................(((((((((((.(((((..............)))))..))))))).))))................... (-20.84 = -21.29 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:42 2011