| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,020,615 – 15,020,708 |
| Length | 93 |
| Max. P | 0.646366 |

| Location | 15,020,615 – 15,020,708 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 77.26 |
| Shannon entropy | 0.47797 |
| G+C content | 0.47014 |
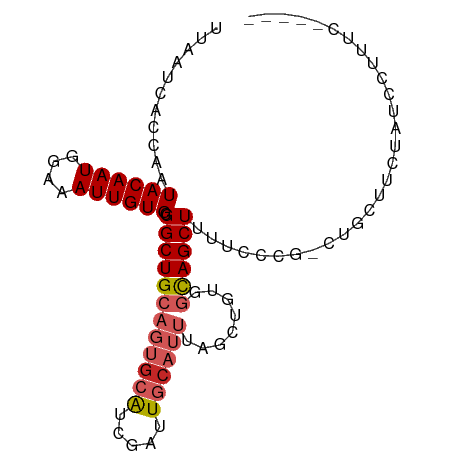
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -12.23 |
| Energy contribution | -13.33 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
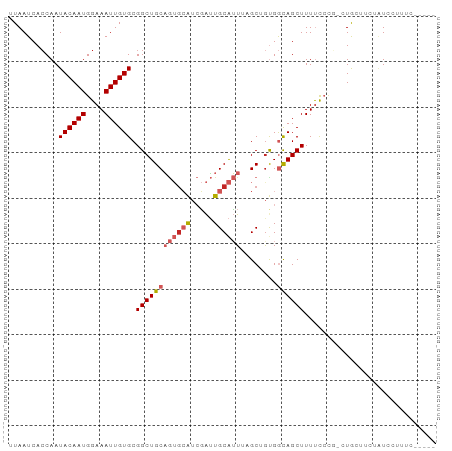
>dm3.chr3L 15020615 93 + 24543557 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCAUCGAUUGCAUUACGCGGUGGCAGCUUUUCCCG-CUGCUUCUAUCCUUUCCGCAA ............(((((....)))))((((((((((((((.....))))))..)))))((((((.......)-)))))...........))).. ( -28.70, z-score = -1.77, R) >droSim1.chr3L 14341759 93 + 22553184 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCAUCGAUUGCAUUUCGCUGUGGCAGCUUUUCCCG-CUGCUUAUAUCCUUUCCACAA ................((((((....(((((.(.((((((.....)))))).))))))((((((.......)-))))).......))))))... ( -26.30, z-score = -1.80, R) >droSec1.super_0 7137380 93 + 21120651 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUACAUCGAUUGCAUUUCGCUGUGGCAGCUUUUCCCG-CUGCUUAUAUCCUUUCCACAA ................((((((....(((((((((((......))))))....)))))((((((.......)-))))).......))))))... ( -24.40, z-score = -1.78, R) >droYak2.chr3L 15111887 93 + 24197627 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCAUCGAUUGCAUUUAGCUGCGGCAGCUUUUCCCG-CUACCUUCAUCCAUUCCACAA ..............((((((......(((((((.((((((.....)))))))))))))((.(((.......)-)).))....))))))...... ( -28.10, z-score = -2.79, R) >droEre2.scaffold_4784 14976080 92 + 25762168 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCAUCGAUUGCAUUGCGCUGUGGCAGCUUUCCCG--CUACUUCCAUCCUUUCCACAA ................((((((..(((.(((.((((((((.....)))))))))))(((((.........)--))))...)))..))))))... ( -27.50, z-score = -2.18, R) >droAna3.scaffold_13337 5329133 80 - 23293914 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCG-CGAUUGCAUUUAGCGACGGCAGCUUUUUCC--CUAGUCCCAU----------- ...........((((((....)))))).((((((.((.((-(...........))))).))))))......--..........----------- ( -19.70, z-score = -0.20, R) >dp4.chrXR_group6 12600924 89 - 13314419 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCAUCGAUUGCAUUCGGCGGUGGCAGCUUUUCCUGGGUGCCUCUGUUGCUGC----- ............(((((....)))))(((((.(.((((((.....)))))))(((((.((((.((......)).)))).))))))))))----- ( -30.90, z-score = -1.16, R) >droPer1.super_12 1017897 89 - 2414086 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCAUCGAUUGCAUUCGGCGGUGGCAGCUUUUCCUGGGUGCCUCUGUUGCUGC----- ............(((((....)))))(((((.(.((((((.....)))))))(((((.((((.((......)).)))).))))))))))----- ( -30.90, z-score = -1.16, R) >droVir3.scaffold_13049 17641437 84 + 25233164 UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCU--GAAUGCGAUGCGGU-AGCAGUGAUAGCUUUCCAUU-GUACUGACUUACUCC------ ........((.(((((((((((((((((.((((((--(.........))))-))).)).)))).))))))))-))).)).........------ ( -26.30, z-score = -2.90, R) >droMoj3.scaffold_6680 22686329 85 + 24764193 UUAAUCAACAAUACAAUGGAAAUUGUGCGGCUGCUA-GAAUGUAGUGCGGU-AGCUGUGAUAGCUUUUCAUU-GUGCCCAGUUGCUCC------ .....((((..((((((((((((((((((((((((.-...........)))-)))))).)))).))))))))-)))....))))....------ ( -24.50, z-score = -1.37, R) >consensus UUAAUCACCAAUACAAUGGAAAUUGUGCGGCUGCAGUGCAUCGAUUGCAUUUAGCUGUGGCAGCUUUUCCCG_CUGCUUCUAUCCUUUC_____ .................(((((...(((.((.((((((((.....))))))..)).)).)))...)))))........................ (-12.23 = -13.33 + 1.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:38 2011