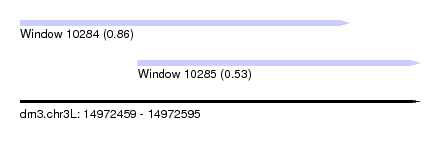
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,972,459 – 14,972,595 |
| Length | 136 |
| Max. P | 0.863301 |

| Location | 14,972,459 – 14,972,571 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.76 |
| Shannon entropy | 0.23042 |
| G+C content | 0.50223 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.863301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
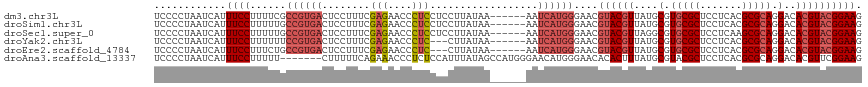
>dm3.chr3L 14972459 112 + 24543557 UCCCCUAAUCAUUUCCUUUUCGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAA------AAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAG ............((((..(((.((((((........(((.......))).......------..))))))))).((((((....(.(((((.......))))).)..)))))))))). ( -30.93, z-score = -2.61, R) >droSim1.chr3L 14289388 112 + 22553184 UCCCCUAAUCAUUUCCUUUUUGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAA------AAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAG ............((((...((.((((((........(((.......))).......------..)))))).)).((((((....(.(((((.......))))).)..)))))))))). ( -30.03, z-score = -2.44, R) >droSec1.super_0 7088993 112 + 21120651 UCCCCUAAUCAUUUCCUUUUUGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAA------AAUCAUGGGAACGUACGUUAGGCGUGCGCUCCUCAAGCGCAGGACACGUACGGAAG ............((((...((.((((((........(((.......))).......------..)))))).)).((((((....(.((((((.....)))))).)..)))))))))). ( -31.23, z-score = -2.14, R) >droYak2.chr3L 15060185 109 + 24197627 UCCCCUAAUCAUUUCCUUUUUUCCGUGACUCCUUUCGAGAACCCUC---CUUAUAA------AAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAG ............((((...(((((((((........(((....)))---.......------..))))))))).((((((....(.(((((.......))))).)..)))))))))). ( -32.13, z-score = -3.56, R) >droEre2.scaffold_4784 14929126 109 + 25762168 UCCCCUAAUCAUUUCCUUUCUGCCGUGACUCCUUUCGAGAACCCUC---CUUAUAA------AAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAG ............((((.(((((((((((........(((....)))---.......------.......(((.((((((.....)))))).)))))))).)))))).......)))). ( -30.00, z-score = -2.43, R) >droAna3.scaffold_13337 19145220 111 - 23293914 UCCCCUAAUCAUUUCCUUUUU-------CUUUUUCAGAAACCCUCUCCAUUUAUAGCCAUGGGAACAUGGGAACACACUUUAUGCGUACGCUCCUCACGCGCAGGACACGUUCGGAAG ............((((..(((-------((.....))))).....(((........(((((....)))))............(((((...........)))))))).......)))). ( -21.20, z-score = -0.64, R) >consensus UCCCCUAAUCAUUUCCUUUUUGCCGUGACUCCUUUCGAGAACCCUCCUCCUUAUAA______AAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAG ............((((............(((.....)))..(((........................)))...((((((....(.(((((.......))))).)..)))))))))). (-20.57 = -21.13 + 0.56)


| Location | 14,972,499 – 14,972,595 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.04 |
| Shannon entropy | 0.42011 |
| G+C content | 0.53189 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -19.36 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.526205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14972499 96 + 24543557 --------ACCCUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCG---AAACCGAAA------ --------.(((.(.((((...........)))).(((((((....(.(((((.......))))).)..))))))).........).)))..(((---....)))..------ ( -28.40, z-score = -1.33, R) >droSim1.chr3L 14289428 96 + 22553184 --------ACCCUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCG---AAACCGAAA------ --------.(((.(.((((...........)))).(((((((....(.(((((.......))))).)..))))))).........).)))..(((---....)))..------ ( -28.40, z-score = -1.33, R) >droSec1.super_0 7089033 96 + 21120651 --------ACCCUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAGGCGUGCGCUCCUCAAGCGCAGGACACGUACGGAAGAAACCGUGGUAUUCG---AAACCGAAA------ --------.................(((((((...(((((((....(.((((((.....)))))).)..))))))).......))))))).((((---....)))).------ ( -30.60, z-score = -2.05, R) >droYak2.chr3L 15060225 92 + 24197627 -----------ACCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCG---AAACCGAA------- -----------.(((((((...........)))).(((((((....(.(((((.......))))).)..)))))))...........)))..(((---....))).------- ( -28.20, z-score = -1.42, R) >droEre2.scaffold_4784 14929166 92 + 25762168 -----------ACCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAAUCGUGGGAAUCG---AAACCGAA------- -----------.(((((((...........)))).(((((((....(.(((((.......))))).)..)))))))...........)))..(((---....))).------- ( -28.20, z-score = -1.47, R) >droAna3.scaffold_13337 19145251 113 - 23293914 AACCCUCUCCAUUUAUAGCCAUGGGAACAUGGGAACACACUUUAUGCGUACGCUCCUCACGCGCAGGACACGUUCGGAAGGAACCGUGGGAAUCGGGGAACCCAAGAAUCGUU ..((((.(((........(((((....)))))...(((.......((....))((((..((.((.......)).))..))))...))))))...))))............... ( -29.80, z-score = 0.53, R) >droPer1.super_12 966076 79 - 2414086 -------------------------AUGGCGAGGACAUGCUCCAUGCGUACGCUGCUCACGCGCAGGACACGCACGGAAGGAACCGUGAGAACCG---AACCCGAAA------ -------------------------....((.((......(((.(((((...((((......))))...))))).))).((...(....)..)).---..))))...------ ( -22.00, z-score = 0.13, R) >consensus _________C_CUCCUCCUUAUAAAAUCAUGGGAACGUACGUUAUGCGUGCGCUCCUCACGCGCAGGACACGUACGGAAGAAACCGUGGGAAUCG___AAACCGAAA______ ..........................((((((...(((((((....(.(((((.......))))).)..))))))).......))))))........................ (-19.36 = -20.24 + 0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:30 2011