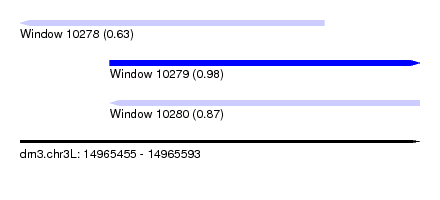
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,965,455 – 14,965,593 |
| Length | 138 |
| Max. P | 0.975988 |

| Location | 14,965,455 – 14,965,560 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.82 |
| Shannon entropy | 0.46991 |
| G+C content | 0.41243 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -12.58 |
| Energy contribution | -12.11 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
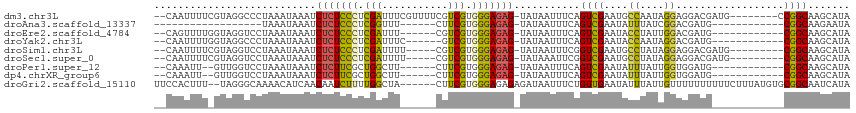
>dm3.chr3L 14965455 105 - 24543557 --CAAUUUUCGUAGGCCCUAAAUAAAUCUCUCCCUCGAUUUCGUUUUCGUCGUGGGAGAG-UAUAAUUUCAGUCGAAUGCCAAUAGGAGGACGAUG--------CCGGCAAGCAUA --........((..(((..........(((((((.((((.........)))).)))))))-..........((((....((....))....)))).--------..)))..))... ( -27.90, z-score = -1.12, R) >droAna3.scaffold_13337 19139016 79 + 23293914 ------------------UAAAUAAAUCUCUCCCUCGGUUU------CUUCGUGGGAGAG-UAUAAUUUCAGUCGAAUAUUUAUCGGACGAUG------------CGGCAAGAAUA ------------------.........(((((((.(((...------..))).)))))))-....((((..((((......((((....))))------------))))..)))). ( -20.60, z-score = -1.62, R) >droEre2.scaffold_4784 14922106 95 - 25762168 --CAGUUUUGGUAGGUCCUAAAUAAAUCUCUCCCUCGAUUU------CGUCGUGGGAGAG-UAUAAUUUCAGUCGAAUACCUAUUGGACGAUG------------CGGCAAGCAUA --..((.(..((((((...........(((((((.((((..------.)))).)))))))-......(((....))).))))))..))).(((------------(.....)))). ( -29.90, z-score = -2.86, R) >droYak2.chr3L 15052993 95 - 24197627 --CAAUUUUGGUAGGCCCUAAAUAAAUCUCUCCCUCGAUUUC------GUCGUGGGAGAG-UAUAAUUUCAGUCGAAUACCAAUAGGACGAUG------------CGGCAAGCAUA --.....((((((..(.((((((....(((((((.((((...------)))).)))))))-....)))).))..)..)))))).......(((------------(.....)))). ( -28.60, z-score = -2.64, R) >droSim1.chr3L 14282168 99 - 22553184 --CAAUUUUCGUAGGUCCUAAAUAAAUCUCUCCCUCGAUUUU-----CGUCGUGGGAGAG-UAUAAUUUCGGUCGAAUGCCUAUAGGAGGACGAUG---------CGGCAAGCAUA --...(((..((((((((.((((....(((((((.((((...-----.)))).)))))))-....)))).))......))))))..)))....(((---------(.....)))). ( -31.40, z-score = -2.37, R) >droSec1.super_0 7081960 99 - 21120651 --CAAUUUUCGUAGGUCCUAAAUAAAUCUCUCCCUCGAUUUU-----CGUCGUGGGAGAG-UAUAAAUUCGGUCGAAUGCCUAUAGGAGGACGAUG---------CGGCAAGCAUA --......(((((.(((((........(((((((.((((...-----.)))).)))))))-((((.((((....))))...))))..)))))..))---------)))........ ( -31.30, z-score = -2.39, R) >droPer1.super_12 959973 93 + 2414086 --CAAAUU--GUUGGUCCUAAAUAAAUCUCUUCGCUGGCUU------CUUCGUGGGAGAG-UAUAAUUUCAGUCGAAUAUUUAUUGGUGGAUG------------CGGCAAGCAUA --....((--((((((((..(((((((...(((((((((((------((....)))))..-.......)))).)))).)))))))...)))).------------))))))..... ( -21.60, z-score = -0.96, R) >dp4.chrXR_group6 12545069 93 + 13314419 --CAAAUU--GUUGGUCCUAAAUAAAUCUCUUCGCUGGCUU------CUUCGUGGGAGAG-UAUAAUUUCAGUCGAAUAUUUAUUGGUGGAUG------------CGGCAAGCAUA --....((--((((((((..(((((((...(((((((((((------((....)))))..-.......)))).)))).)))))))...)))).------------))))))..... ( -21.60, z-score = -0.96, R) >droGri2.scaffold_15110 6090036 108 + 24565398 UUCCACUUU--UAGGGCAAAACAUCAACAAUCUUUUGGCUA------CUUCGUGGGAGAGAGAUAAUUUCUGUUGAAUAUUUAUUGUUUUUUUUUUCUUUAUGUGCGGCAAUCAUA .(((((...--.((((.(((((((((((((((((((..(((------(...))))..)))))))......))))))........))))))....))))....))).))........ ( -16.60, z-score = 0.47, R) >consensus __CAAUUUU_GUAGGUCCUAAAUAAAUCUCUCCCUCGAUUU______CGUCGUGGGAGAG_UAUAAUUUCAGUCGAAUACCUAUAGGAGGAUG____________CGGCAAGCAUA ...........................(((((((.(((...........))).)))))))...........((((..............................))))....... (-12.58 = -12.11 + -0.47)
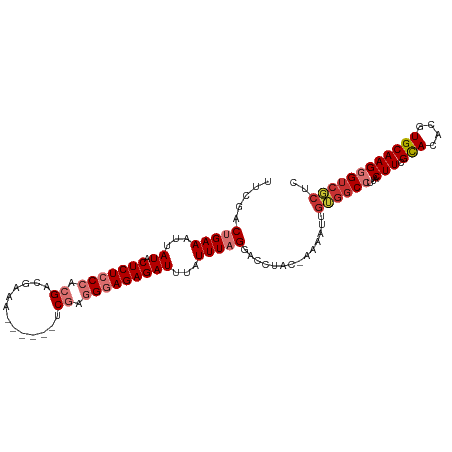
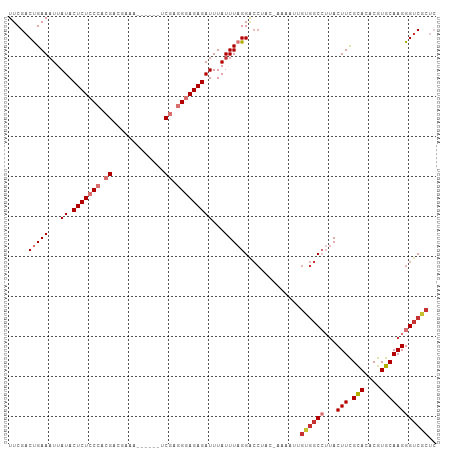
| Location | 14,965,486 – 14,965,593 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.51 |
| Shannon entropy | 0.29892 |
| G+C content | 0.45967 |
| Mean single sequence MFE | -28.59 |
| Consensus MFE | -20.87 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
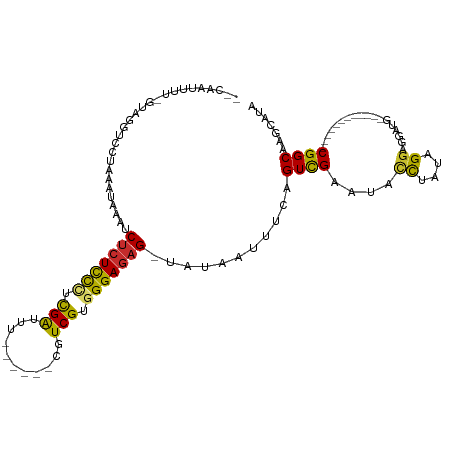
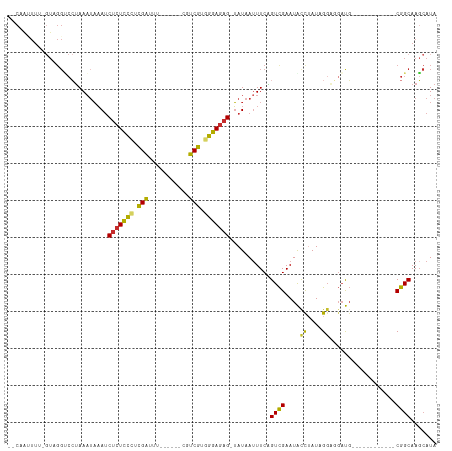
>dm3.chr3L 14965486 107 + 24543557 UUCGACUGAAAUUAUACUCUCCCACGACGAAAACGAAAUCGAGGGAGAGAUUUAUUUAGGGCCUACGAAAAUUGUGGCCUUACUUCGCACACGUGCAAGGGUCGCUC ..((((((((......(((((((.(((((....))...))).))))))).......(((((((.((((...))))))))))).)))(((....)))...)))))... ( -36.80, z-score = -4.04, R) >droAna3.scaffold_13337 19139043 84 - 23293914 UUCGACUGAAAUUAUACUCUCCCACGAAGAAA------CCGAGGGAGAGAUUUAUUUAGG-----------------CCUCACUUUGUACACGUGCAAGGGACUCUC .....(((((...((.(((((((.((......------.)).)))))))))...))))).-----------------..((.(((((((....)))))))))..... ( -24.40, z-score = -2.44, R) >droEre2.scaffold_4784 14922133 101 + 25762168 UUCGACUGAAAUUAUACUCUCCCACGACGAAA------UCGAGGGAGAGAUUUAUUUAGGACCUACCAAAACUGUGGCCUUACUUCGCACACGUGCAAGGGUCGCUC .....(((((...((.(((((((.(((.....------))).)))))))))...)))))(((((.(((......))).........(((....)))..))))).... ( -30.80, z-score = -2.94, R) >droYak2.chr3L 15053020 101 + 24197627 UUCGACUGAAAUUAUACUCUCCCACGACGAAA------UCGAGGGAGAGAUUUAUUUAGGGCCUACCAAAAUUGUGGCCUUACUUCGCACACGUGCAAGGGUCGCUC ..((((((((......(((((((.(((.....------))).))))))).......(((((((.((.......))))))))).)))(((....)))...)))))... ( -35.50, z-score = -4.02, R) >droSim1.chr3L 14282198 102 + 22553184 UUCGACCGAAAUUAUACUCUCCCACGACGAAAA-----UCGAGGGAGAGAUUUAUUUAGGACCUACGAAAAUUGUGGCCUUACUUCGCACACGUGCAAGGGUCGCUC ..((((((((......(((((((.(((......-----))).)))))))........(((.((.((((...)))))))))...)))(((....)))...)))))... ( -33.80, z-score = -3.53, R) >droSec1.super_0 7081990 102 + 21120651 UUCGACCGAAUUUAUACUCUCCCACGACGAAAA-----UCGAGGGAGAGAUUUAUUUAGGACCUACGAAAAUUGUGGCCUUACUUCGCACACGUGCAAGGGUCGCUC ..(((((((((..((.(((((((.(((......-----))).)))))))))..))))(((.((.((((...)))))))))..(((.(((....)))))))))))... ( -34.20, z-score = -3.63, R) >droPer1.super_12 960000 97 - 2414086 UUCGACUGAAAUUAUACUCUCCCACGAAGAAG------CCAGCGAAGAGAUUUAUUUAGGACCAAC--AAUUUGAGGCAAUACUUUGCACACGUGCAAGGGUCUC-- .....(((((...((.((((....((..(...------.)..)).))))))...))))).......--.....(((((....(((((((....))))))))))))-- ( -16.60, z-score = 0.43, R) >dp4.chrXR_group6 12545096 97 - 13314419 UUCGACUGAAAUUAUACUCUCCCACGAAGAAG------CCAGCGAAGAGAUUUAUUUAGGACCAAC--AAUUUGAGGCAAUACUUUGCACACGUGCAAGGGUCUC-- .....(((((...((.((((....((..(...------.)..)).))))))...))))).......--.....(((((....(((((((....))))))))))))-- ( -16.60, z-score = 0.43, R) >consensus UUCGACUGAAAUUAUACUCUCCCACGACGAAA______UCGAGGGAGAGAUUUAUUUAGGACCUAC_AAAAUUGUGGCCUUACUUCGCACACGUGCAAGGGUCGCUC .....(((((...((.(((((((.((.............)).)))))))))...)))))..............((((((...(((.(((....)))))))))))).. (-20.87 = -22.20 + 1.33)

| Location | 14,965,486 – 14,965,593 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.51 |
| Shannon entropy | 0.29892 |
| G+C content | 0.45967 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -20.21 |
| Energy contribution | -20.15 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
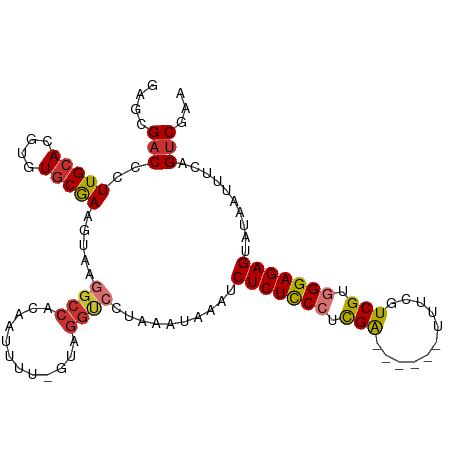
>dm3.chr3L 14965486 107 - 24543557 GAGCGACCCUUGCACGUGUGCGAAGUAAGGCCACAAUUUUCGUAGGCCCUAAAUAAAUCUCUCCCUCGAUUUCGUUUUCGUCGUGGGAGAGUAUAAUUUCAGUCGAA ...((((....(((....)))(((((..((((((.......)).))))..........(((((((.((((.........)))).)))))))....))))).)))).. ( -34.60, z-score = -3.24, R) >droAna3.scaffold_13337 19139043 84 + 23293914 GAGAGUCCCUUGCACGUGUACAAAGUGAGG-----------------CCUAAAUAAAUCUCUCCCUCGG------UUUCUUCGUGGGAGAGUAUAAUUUCAGUCGAA ((((...(((..(...........)..)))-----------------...........(((((((.(((------.....))).))))))).....))))....... ( -22.90, z-score = -1.67, R) >droEre2.scaffold_4784 14922133 101 - 25762168 GAGCGACCCUUGCACGUGUGCGAAGUAAGGCCACAGUUUUGGUAGGUCCUAAAUAAAUCUCUCCCUCGA------UUUCGUCGUGGGAGAGUAUAAUUUCAGUCGAA ...((((....(((....)))(((((.(((((((.......)).)).)))........(((((((.(((------(...)))).)))))))....))))).)))).. ( -32.40, z-score = -2.59, R) >droYak2.chr3L 15053020 101 - 24197627 GAGCGACCCUUGCACGUGUGCGAAGUAAGGCCACAAUUUUGGUAGGCCCUAAAUAAAUCUCUCCCUCGA------UUUCGUCGUGGGAGAGUAUAAUUUCAGUCGAA ...((((....(((....)))(((((..((((((.......)).))))..........(((((((.(((------(...)))).)))))))....))))).)))).. ( -34.60, z-score = -3.35, R) >droSim1.chr3L 14282198 102 - 22553184 GAGCGACCCUUGCACGUGUGCGAAGUAAGGCCACAAUUUUCGUAGGUCCUAAAUAAAUCUCUCCCUCGA-----UUUUCGUCGUGGGAGAGUAUAAUUUCGGUCGAA ...(((((...(((....)))(((((.(((((((.......)).)).)))........(((((((.(((-----(....)))).)))))))....)))))))))).. ( -36.60, z-score = -3.78, R) >droSec1.super_0 7081990 102 - 21120651 GAGCGACCCUUGCACGUGUGCGAAGUAAGGCCACAAUUUUCGUAGGUCCUAAAUAAAUCUCUCCCUCGA-----UUUUCGUCGUGGGAGAGUAUAAAUUCGGUCGAA ...(((((...(((....)))(((...(((((((.......)).)).)))........(((((((.(((-----(....)))).)))))))......)))))))).. ( -34.70, z-score = -3.25, R) >droPer1.super_12 960000 97 + 2414086 --GAGACCCUUGCACGUGUGCAAAGUAUUGCCUCAAAUU--GUUGGUCCUAAAUAAAUCUCUUCGCUGG------CUUCUUCGUGGGAGAGUAUAAUUUCAGUCGAA --..((((...(((..((.((((....))))..))...)--)).))))..............(((((((------(((((....))))).........)))).))). ( -17.70, z-score = 0.90, R) >dp4.chrXR_group6 12545096 97 + 13314419 --GAGACCCUUGCACGUGUGCAAAGUAUUGCCUCAAAUU--GUUGGUCCUAAAUAAAUCUCUUCGCUGG------CUUCUUCGUGGGAGAGUAUAAUUUCAGUCGAA --..((((...(((..((.((((....))))..))...)--)).))))..............(((((((------(((((....))))).........)))).))). ( -17.70, z-score = 0.90, R) >consensus GAGCGACCCUUGCACGUGUGCGAAGUAAGGCCACAAUUUU_GUAGGUCCUAAAUAAAUCUCUCCCUCGA______UUUCGUCGUGGGAGAGUAUAAUUUCAGUCGAA ....(((..(((((....))))).....((((............))))..........(((((((.(((...........))).)))))))..........)))... (-20.21 = -20.15 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:26 2011