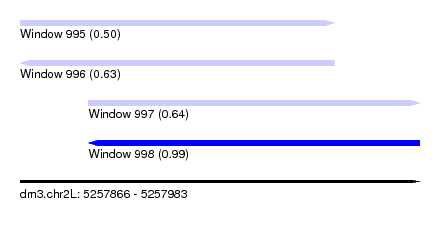
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,257,866 – 5,257,983 |
| Length | 117 |
| Max. P | 0.994635 |

| Location | 5,257,866 – 5,257,958 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.47743 |
| G+C content | 0.40008 |
| Mean single sequence MFE | -18.98 |
| Consensus MFE | -8.41 |
| Energy contribution | -8.54 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
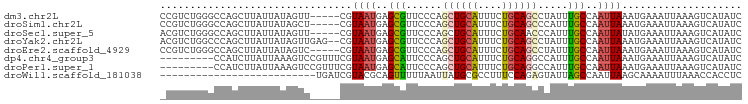
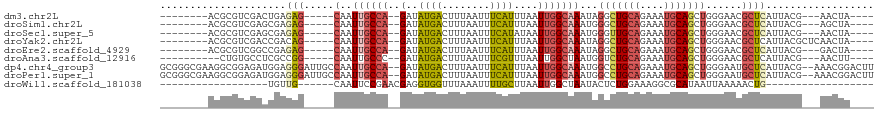
>dm3.chr2L 5257866 92 + 23011544 CCGUCUGGGCCAGCUUAUUAUAGUU-----CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC .....((((...(((((((((....-----.)))))))))...))))((((((....))))))...............((((........))))... ( -25.00, z-score = -2.99, R) >droSim1.chr2L 5056382 92 + 22036055 CCGUCUGGGCCAGCUUAUUAUAGCU-----CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC .....((((...(((((((((....-----.)))))))))...))))((((((....)))))).((((((.....))))))................ ( -26.20, z-score = -3.11, R) >droSec1.super_5 3338064 92 + 5866729 ACGUCUGGGCCAGCUUAUUAUAGUU-----CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAACCCAUUUGCCAAUUAUAUGAAAUUAAAGUCAUAUC ..(.(((((...(((((((((....-----.)))))))))...))))))((((....))))...............((((((........)))))). ( -23.40, z-score = -2.77, R) >droYak2.chr2L 8383585 95 - 22324452 ACGUCUGGCCCAGCUUAUUAUAGUUGAG--CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC .....((((...(((((((((.......--.))))))))).......((((((....))))))......)))).....((((........))))... ( -24.10, z-score = -2.16, R) >droEre2.scaffold_4929 5336494 92 + 26641161 CCGUCUGGGCCAGCUUAUUAUAGUC-----CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC .....((((...(((((((((....-----.)))))))))...))))((((((....))))))...............((((........))))... ( -25.00, z-score = -3.06, R) >dp4.chr4_group3 7054216 88 + 11692001 ---------CCAUCUUAUUAAAGUCCGUUUCGUAAUGAGCAUUCCCAGCUGCAUUUCUGCAGGCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC ---------.................((((((((((..(((.......(((((....)))))......)))..)))..)))))))............ ( -12.22, z-score = -0.28, R) >droPer1.super_1 8506565 88 + 10282868 ---------CCAUCUUAUUAAAGUCCGUUUCGUAAUGAGCAUUCCCAGCUGCAUUUCUGCAGGCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC ---------.................((((((((((..(((.......(((((....)))))......)))..)))..)))))))............ ( -12.22, z-score = -0.28, R) >droWil1.scaffold_181038 186216 71 + 637489 --------------------------UGAUCGUACGCAGUUUUUAAUUAUGCGCCUUUCCAGAGUAUUAGCCAAUUAAGCAAAAUUUAAACCACCUC --------------------------........((((...........))))................((.......))................. ( -3.70, z-score = 1.50, R) >consensus _CGUCUGGGCCAGCUUAUUAUAGUU_____CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC ................................((((..(((......((((((....)))))).....)))..)))).................... ( -8.41 = -8.54 + 0.13)

| Location | 5,257,866 – 5,257,958 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.47743 |
| G+C content | 0.40008 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -11.96 |
| Energy contribution | -12.41 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.628149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5257866 92 - 23011544 GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG-----AACUAUAAUAAGCUGGCCCAGACGG .................................(((((((....)))))))(((...(((.((((..-----.....)))).)))...)))...... ( -21.90, z-score = -1.61, R) >droSim1.chr2L 5056382 92 - 22036055 GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG-----AGCUAUAAUAAGCUGGCCCAGACGG ...............................(((((((((....)))(((((....)((((.....)-----))).......))))))))))..... ( -24.60, z-score = -1.66, R) >droSec1.super_5 3338064 92 - 5866729 GAUAUGACUUUAAUUUCAUAUAAUUGGCAAAUGGGUUGCAGAAAUGCAGCUGGGAACGCUCAUUACG-----AACUAUAAUAAGCUGGCCCAGACGU .((((((........))))))............(((((((....)))))))(((...(((.((((..-----.....)))).)))...)))...... ( -22.80, z-score = -1.72, R) >droYak2.chr2L 8383585 95 + 22324452 GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG--CUCAACUAUAAUAAGCUGGGCCAGACGU ....................((((.(((.....(((((((....)))))))(....)))).)))).(--((((.((......)).)))))....... ( -24.20, z-score = -1.62, R) >droEre2.scaffold_4929 5336494 92 - 26641161 GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG-----GACUAUAAUAAGCUGGCCCAGACGG .................................(((((((....)))))))(((...(((.((((..-----.....)))).)))...)))...... ( -21.90, z-score = -1.38, R) >dp4.chr4_group3 7054216 88 - 11692001 GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAAUGCUCAUUACGAAACGGACUUUAAUAAGAUGG--------- ....(((..((..((((...((((.((((..((((.((((....))))))))....)))).)))).))))..))..))).........--------- ( -15.80, z-score = -0.51, R) >droPer1.super_1 8506565 88 - 10282868 GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAAUGCUCAUUACGAAACGGACUUUAAUAAGAUGG--------- ....(((..((..((((...((((.((((..((((.((((....))))))))....)))).)))).))))..))..))).........--------- ( -15.80, z-score = -0.51, R) >droWil1.scaffold_181038 186216 71 - 637489 GAGGUGGUUUAAAUUUUGCUUAAUUGGCUAAUACUCUGGAAAGGCGCAUAAUUAAAAACUGCGUACGAUCA-------------------------- (((.((((...((((......)))).))))...)))..((...(((((...........)))))....)).-------------------------- ( -10.60, z-score = 0.37, R) >consensus GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG_____AACUAUAAUAAGCUGGCCCAGACG_ ...((((........)))).((((.(((...(.(((((((....))))))).)....))).))))................................ (-11.96 = -12.41 + 0.45)
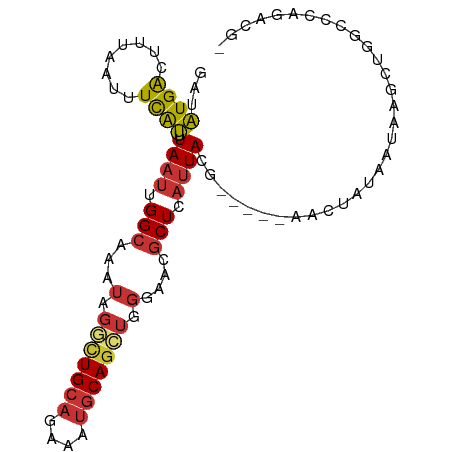
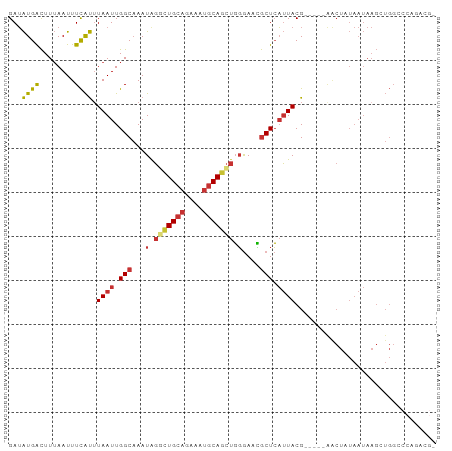
| Location | 5,257,886 – 5,257,983 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.83 |
| Shannon entropy | 0.50331 |
| G+C content | 0.44098 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -11.09 |
| Energy contribution | -12.53 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
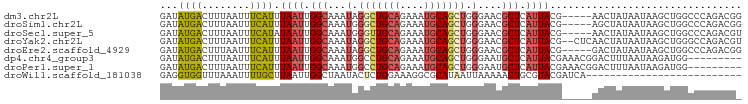
>dm3.chr2L 5257886 97 + 23011544 ----UAGUU---CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC--UGGCAAUUG-----CUCUCAGUCGACGCGU-------- ----..(((---((....(((((......((((((....))))))....((((((.....((((........))))...--)))))).))-----))).....))).))..-------- ( -26.00, z-score = -2.37, R) >droSim1.chr2L 5056402 97 + 22036055 ----UAGCU---CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC--UGGCAAUUG-----CUCUCGCUCGACGCGU-------- ----..((.---(((...(((((......((((((....)))))).((.((((((.....((((........))))...--)))))).))-----....))))).))).))-------- ( -28.80, z-score = -2.79, R) >droSec1.super_5 3338084 97 + 5866729 ----UAGUU---CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAACCCAUUUGCCAAUUAUAUGAAAUUAAAGUCAUAUC--UGGCAAUUG-----CUCUCGCUCGACGCGU-------- ----.....---(((..((((((.....(((((((....))))......((((((...((((((........)))))).--))))))..)-----))..))))))..))).-------- ( -26.30, z-score = -2.72, R) >droYak2.chr2L 8383605 100 - 22324452 ----UAGUUGAGCGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC--UGGCAAUUG-----CUGUCGGUCGACGCGU-------- ----..(((((.((.....((((......((((((....))))))....((((((.....((((........))))...--)))))).))-----))..)).)))))....-------- ( -28.00, z-score = -1.43, R) >droEre2.scaffold_4929 5336514 97 + 26641161 ----UAGUC---CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCUAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC--UGGCAAUUG-----CUCUCGGCCGACGCGU-------- ----..(((---.((...(((((......((((((....))))))....((((((.....((((........))))...--)))))).))-----)))...)).)))....-------- ( -29.20, z-score = -2.82, R) >droAna3.scaffold_12916 6274220 95 - 16180835 ----AAGUU---CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGACCAUUAGCCAAUUAAACGAAAUUAAAGUCAUAUC--GGGCAAUUG-----CCGGCGAGGCACAG---------- ----..(((---(.....))))((.(((..(((((....))))).......(((.......(((.((.......)).))--)(((....)-----)))))).)).))..---------- ( -22.70, z-score = -0.64, R) >dp4.chr4_group3 7054227 115 + 11692001 AAGUCCGUUU--CGUAAUGAGCAUUCCCAGCUGCAUUUCUGCAGGCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC--UGGCAAUUGGCAAUCCCUCCAUCUCCGCCUUCGCCCGC ..(..((((.--...))))..)........(((((....)))))((((.((((((.....((((........))))...--)))))).))))........................... ( -25.40, z-score = -1.19, R) >droPer1.super_1 8506576 115 + 10282868 AAGUCCGUUU--CGUAAUGAGCAUUCCCAGCUGCAUUUCUGCAGGCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC--UGGCAAUUGGCAAUCCCUCCAUCUCCGCCUUCGCCCGC ..(..((((.--...))))..)........(((((....)))))((((.((((((.....((((........))))...--)))))).))))........................... ( -25.40, z-score = -1.19, R) >droWil1.scaffold_181038 186226 77 + 637489 ------------------CAGUUUUUAAUUAUGCGCCUUUCCAGAGUAUUAGCCAAUUAAGCAAAAUUUAAACCACCUCGUUCGGAAUUG------CAACA------------------ ------------------................((..((((.(((........((((......))))........)))....))))..)------)....------------------ ( -7.59, z-score = 0.56, R) >consensus ____UAGUU___CGUAAUGAGCGUUCCCAGCUGCAUUUCUGCAGCCCAUUUGCCAAUUAAAUGAAAUUAAAGUCAUAUC__UGGCAAUUG_____CUCUCGAUCGACGCGU________ ....................((.......((((((....)))))).((.((((((.....((((........)))).....)))))).)).................)).......... (-11.09 = -12.53 + 1.45)

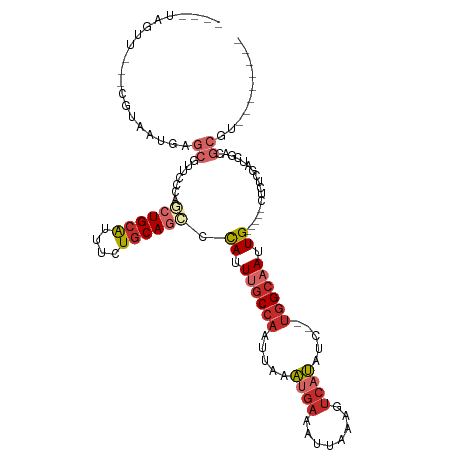
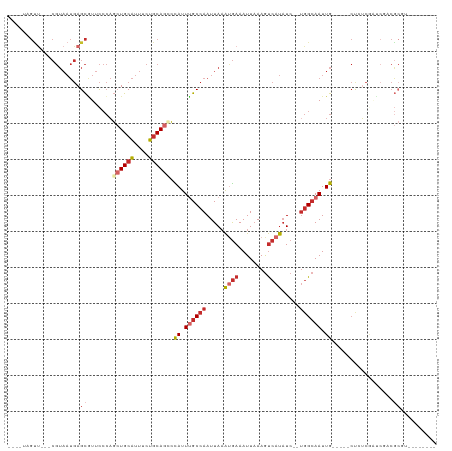
| Location | 5,257,886 – 5,257,983 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.83 |
| Shannon entropy | 0.50331 |
| G+C content | 0.44098 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -17.82 |
| Energy contribution | -18.59 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.994635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
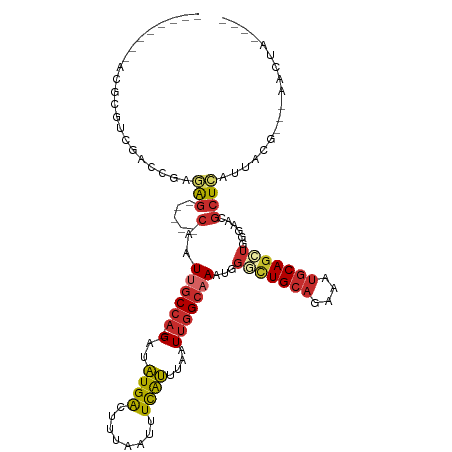
>dm3.chr2L 5257886 97 - 23011544 --------ACGCGUCGACUGAGAG-----CAAUUGCCA--GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG---AACUA---- --------.....(((..((((((-----(..((((((--(..((((........))))....)))))))...(((((((....)))))))......)))).)))))---)....---- ( -30.60, z-score = -3.47, R) >droSim1.chr2L 5056402 97 - 22036055 --------ACGCGUCGAGCGAGAG-----CAAUUGCCA--GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG---AGCUA---- --------.(((.....))).(((-----(..((((((--(..((((........))))....))))))).(.(((((((....))))))).)....))))......---.....---- ( -34.40, z-score = -3.38, R) >droSec1.super_5 3338084 97 - 5866729 --------ACGCGUCGAGCGAGAG-----CAAUUGCCA--GAUAUGACUUUAAUUUCAUAUAAUUGGCAAAUGGGUUGCAGAAAUGCAGCUGGGAACGCUCAUUACG---AACUA---- --------.(((.....))).(((-----(..((((((--.((((((........))))))...)))))).(.(((((((....))))))).)....))))......---.....---- ( -34.50, z-score = -4.32, R) >droYak2.chr2L 8383605 100 + 22324452 --------ACGCGUCGACCGACAG-----CAAUUGCCA--GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACGCUCAACUA---- --------..(((((....)))((-----(..((((((--(..((((........))))....)))))))...(((((((....)))))))......)))......)).......---- ( -30.80, z-score = -3.17, R) >droEre2.scaffold_4929 5336514 97 - 26641161 --------ACGCGUCGGCCGAGAG-----CAAUUGCCA--GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUAGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG---GACUA---- --------....(((.(....(((-----(..((((((--(..((((........))))....)))))))...(((((((....)))))))......))))....).---)))..---- ( -33.20, z-score = -3.42, R) >droAna3.scaffold_12916 6274220 95 + 16180835 ----------CUGUGCCUCGCCGG-----CAAUUGCCC--GAUAUGACUUUAAUUUCGUUUAAUUGGCUAAUGGUCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG---AACUU---- ----------.((.((.((.(((.-----((((((..(--((.((.......)).)))..)))))).)....((.(((((....)))))))))))..)).)).....---.....---- ( -23.50, z-score = -0.42, R) >dp4.chr4_group3 7054227 115 - 11692001 GCGGGCGAAGGCGGAGAUGGAGGGAUUGCCAAUUGCCA--GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAAUGCUCAUUACG--AAACGGACUU .((.((....)).......(((.(...((((.((((((--(..((((........))))....))))))).))))(((((....))))).......).)))......--...))..... ( -32.60, z-score = -1.25, R) >droPer1.super_1 8506576 115 - 10282868 GCGGGCGAAGGCGGAGAUGGAGGGAUUGCCAAUUGCCA--GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGCCUGCAGAAAUGCAGCUGGGAAUGCUCAUUACG--AAACGGACUU .((.((....)).......(((.(...((((.((((((--(..((((........))))....))))))).))))(((((....))))).......).)))......--...))..... ( -32.60, z-score = -1.25, R) >droWil1.scaffold_181038 186226 77 - 637489 ------------------UGUUG------CAAUUCCGAACGAGGUGGUUUAAAUUUUGCUUAAUUGGCUAAUACUCUGGAAAGGCGCAUAAUUAAAAACUG------------------ ------------------(((.(------(..(((((...(((.((((...((((......)))).))))...))))))))..))))).............------------------ ( -13.20, z-score = -0.18, R) >consensus ________ACGCGUCGACCGAGAG_____CAAUUGCCA__GAUAUGACUUUAAUUUCAUUUAAUUGGCAAAUGGGCUGCAGAAAUGCAGCUGGGAACGCUCAUUACG___AACUA____ ..........(((...................((((((.....((((........)))).....))))))...(((((((....))))))).....))).................... (-17.82 = -18.59 + 0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:35 2011