| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,958,176 – 14,958,302 |
| Length | 126 |
| Max. P | 0.929166 |

| Location | 14,958,176 – 14,958,302 |
|---|---|
| Length | 126 |
| Sequences | 10 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 72.39 |
| Shannon entropy | 0.60026 |
| G+C content | 0.38835 |
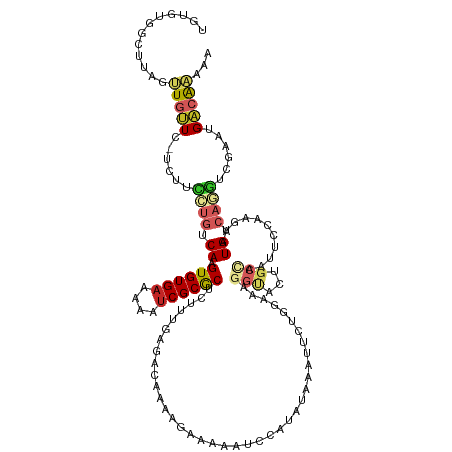
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -10.37 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.929166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
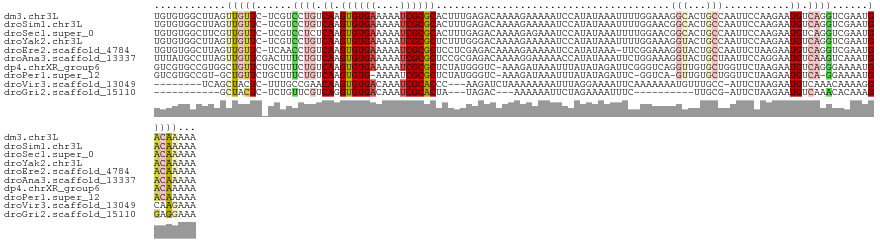
>dm3.chr3L 14958176 126 - 24543557 UGUGUGGCUUAGUUGUUC-UCGUCCUGUCAAGUGUGAAAAAUCGCGCACUUUGAGACAAAAGAAAAAUCCAUAUAAAUUUUGGAAAGGCACUGCCAAUUCCAAGAAUGUCAGGUCGAAUGACAAAAA ............(((((.-(((.((((.((.((((((....)))))).((((......))))...............((((((((.(((...)))..)))))))).)).)))).)))..)))))... ( -32.90, z-score = -2.30, R) >droSim1.chr3L 14274894 126 - 22553184 UGUGUGGCUUAGUUGUUC-UCGUCCUGUCAAGUGUGAAAAAUCGCGCACUUUGAGACAAAAGAAAAAUCCAUAUAAAUUUUGGAACGGCACUGCCAAUUCCAAGAAUGUCAGGUCGAAUGACAAAAA ............(((((.-(((.((((.((.((((((....)))))).((((......))))...............((((((((.(((...)))..)))))))).)).)))).)))..)))))... ( -33.00, z-score = -2.08, R) >droSec1.super_0 7074790 126 - 21120651 UGUGUGGCUUCGUUGUUC-UCGUCCUCUCAAGUGUGAAAAAUCGCGCACUUUGAGACAAAAGAGAAAUCCAUAUAAAUUUUGGAACGGCACUGCCAAUUCCAAGAAUGUCAGGUCGAAUGACAAAAA (((((((........(((-((....(((((((((((........))))).)))))).....)))))..)))))))..((((((((.(((...)))..)))))))).(((((.......))))).... ( -37.40, z-score = -2.91, R) >droYak2.chr3L 15045735 126 - 24197627 UGUGUGGCUUAGUUGUUC-UCGUCCUGUCAAGUGUGAAAAAUCGCGCUCUUUGGGACAAAAGAAAAAUCCAUAUAAAUUUUGGAAAGGUACUGCCAAUUCCAAGAAUGUCAGGUCGAAUGACAAAAA (((((((....(((.(((-(.((((((...(((((((....)))))))...))))))...)))).))))))))))..((((((((.((.....))..)))))))).(((((.......))))).... ( -36.40, z-score = -3.52, R) >droEre2.scaffold_4784 14915497 125 - 25762168 UGUGUGGCUUAGUUGUUC-UCAACCUGUCAAGUGUGAAAAAUCGCGCUCCUCGAGACAAAAGAAAAAUCCAUAUAAA-UUCGGAAAGGUACUGCCAAUUCUAAGAAUGUCAGGUCGAAUGACAAAAA ...........((..(((-...(((((.(((((((((....)))))))............((((...(((.......-...)))..((.....))..)))).....)).))))).)))..))..... ( -26.90, z-score = -1.23, R) >droAna3.scaffold_13337 19132467 127 + 23293914 UUUAUGCCUUAGUUGUUCGACUUUCUGUCAAGUGUGAAAAAUCGCGCUCCGCGAGACAAAAGGAAAAACCAUAUAAAUUCUGGAAAGGUACUGCUAAUUCCAGGAAUGUCAAGUCAAAUGACAAAAA ...........((..((.(((((..(((..(((((((....)))))))..))).((((...((.....)).......((((((((.((.....))..)))))))).))))))))).))..))..... ( -32.60, z-score = -2.74, R) >dp4.chrXR_group6 12538577 126 + 13314419 GUCGUGCCGUGGCUGUUCUGCUUUCUGUCAAGUGUGAAAAAUCGCGCUCUAUGGGUC-AAAGAUAAAUUUAUAUAGAUUCGGGUCAGGUUGUGCUGGUUCUAAGAAUGUCAGGGAAAAUGACAAAAA (((((.((.((((.(((((....(((((..(((((((....)))))))..)))))..-......................(..((((......))))..)..))))))))).))...)))))..... ( -34.90, z-score = -2.35, R) >droPer1.super_12 953454 121 + 2414086 GUCGUGCCGU-GCUGUUCUGCUUUCUGUCAAGUGUG-AAAAUCGCGCUCUAUGGGUC-AAAGAUAAAUUUAUAUAGAUUC-GGUCA-GUUGUGCUGGUUCUAAGAAUGUCA-GGAAAAUGACAAAAA (((((.((.(-(.((((((....(((((..((((((-.....))))))..)))))..-...............((((..(-(((..-.....))))..)))))))))).))-))...)))))..... ( -28.70, z-score = -1.18, R) >droVir3.scaffold_13049 7457839 114 + 25233164 --------UCAGCUACUC-UUUGCCGAACAAGUGUGACAAAUCGCACCC---AAGAUCUAAAAAAAAUUUAGGAAAAUUCAAAAAAAUGUUUGCC-AUUCUAAGAAUGUCAAACAAAAGGCAAGAAA --------........((-((.((((((...((((((....))))))..---....((((((.....))))))....))).......((((((.(-((((...))))).))))))...))))))).. ( -26.40, z-score = -3.74, R) >droGri2.scaffold_15110 6083567 98 + 24565398 -----------GCUACUC-UCUGUUCGUCAGGUGUGACAAAUCGCACUA---UAGAC---AAAAAAUUCUAGAAAAUUUC----------UUGCG-AUUCUAAGAAUGUCAAACACAAAGGAGGAAA -----------....(((-((((.....)))(((((((((((((((...---((((.---.......))))((.....))----------.))))-))).......))))..))))...)))).... ( -18.61, z-score = -0.74, R) >consensus UGUGUGGCUUAGUUGUUC_UCUUCCUGUCAAGUGUGAAAAAUCGCGCUCUUUGAGACAAAAGAAAAAUCCAUAUAAAUUCUGGAAAGGUACUGCCAAUUCCAAGAAUGUCAGGUCGAAUGACAAAAA ............(((((......((((.((.((((((....)))))).......................................((.....))...........)).))))......)))))... (-10.37 = -11.12 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:22 2011