
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,946,516 – 14,946,648 |
| Length | 132 |
| Max. P | 0.666158 |

| Location | 14,946,516 – 14,946,634 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.50 |
| Shannon entropy | 0.63663 |
| G+C content | 0.50798 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -8.89 |
| Energy contribution | -8.47 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
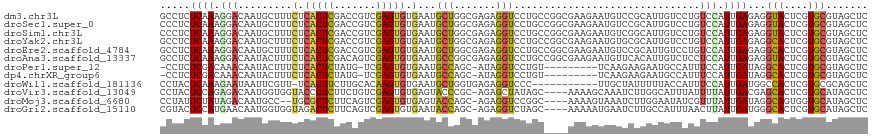
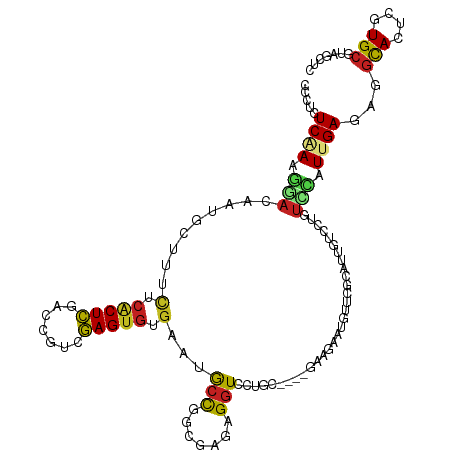
>dm3.chr3L 14946516 118 + 24543557 GCCUCUCAAAGGACAAUGCUUUCUCACUCGACCGUCGAGUGUGAAUGCUGGCGAGAGGUCCUGCCGGCGAAGAAUGUCCGCAUUGUCCUGUCCAUUGAGAGGUACUCGUGCGUAGCUC (((((((((((((((((((.(((.((((((.....)))))).)))((((((((.(....).))))))))..........)))))))))).....)))))))))............... ( -53.50, z-score = -4.73, R) >droSec1.super_0 7064919 118 + 21120651 CCCUCUCAAAGGACAAUGCUUUCUCACUCGACCGUCGAGUGUGAAUGCUGGCGAGAGGUCCUGCCGGCGAAGAAUGUCCGCAUUGUCCUGUCCAUUGAGAGGUACUCGUGCGUAGCUC .((((((((((((((((((.(((.((((((.....)))))).)))((((((((.(....).))))))))..........)))))))))).....))))))))................ ( -51.70, z-score = -4.52, R) >droSim1.chr3L 14264822 118 + 22553184 CCCUCUCAAAGGACAAUGCUUUCUCACUCGACCGUCGAGUGUGAAUGCUGGCGAGAGGUCCUGCCGGCGAAGAAUGUCGGCAUUGUCCUGUCCAUUGAGAGGUACUCGUGCGUAGCUC .((((((((((((((((((((((.((((((.....)))))).)))((((((((.(....).)))))))).........))))))))))).....))))))))................ ( -52.20, z-score = -4.27, R) >droYak2.chr3L 15035214 118 + 24197627 GCCUCUCAAAGGACAAUGCUUUCUCACUCGACCGUCGAGUGUGAAUGCUGGCGAGAGGUCCUGCCGGCGAAGAAUGUGCGCAUUGUCCUGUCCAUUGAGAGGCACUCGUGCGUAGCUC (((((((((((((((((((.(((.((((((.....)))))).)))((((((((.(....).))))))))..........)))))))))).....)))))))))............... ( -55.90, z-score = -4.82, R) >droEre2.scaffold_4784 14905606 118 + 25762168 GCCUCUCAAAGGACAAUGCUUUCUCACUCGACCGUCGAGUGUGAAUGCUGGCGAGAGGUCCUGCCGGCGAAGAAUGUCCGCAUUGUCCUGUCCAUUGAGAGGCACUCGUGCGUAGCUC (((((((((((((((((((.(((.((((((.....)))))).)))((((((((.(....).))))))))..........)))))))))).....)))))))))............... ( -55.90, z-score = -5.09, R) >droAna3.scaffold_13337 19123900 118 - 23293914 GCCUCUCAAAGGACAAUACUUUCUCACUCGACAGUCGAGUGUGAAUGCCGGCGAGAGGUCCUGCCGGCGAAGAAUGUUCACAUUGUCUCCUCCAUUGAGAGGUACUCGUGCGUAGCUC (((((((((.(((.......(((.((((((.....)))))).))).(((((((.(....).)))))))((..((((....)))).))...))).)))))))))............... ( -45.60, z-score = -3.00, R) >droPer1.super_12 943249 106 - 2414086 -CCUCUCGACAAACAAUACUUUCUCACUCUAUG-UCGAGUGUGAAUGCCAGC-AUAGGUCCUGU---------UCAAGAAGAAUGCCAUUUCCAUUGAUAGGCACUCGUGCGUAGCUC -...................(((.(((((....-..))))).))).((..((-(..((((((((---------.(((...(((......)))..)))))))).)))..)))...)).. ( -27.10, z-score = -1.47, R) >dp4.chrXR_group6 12526120 106 - 13314419 -CCUCUCGACAAACAAUACUUUCUCACUCUAUG-UCGAGUGUGAAUGCCAGC-AUAGGUCCUGU---------UCAAGAAGAAUGCCAUUUCCAUUGAUAGGCACUCGUGCGUAGCUC -...................(((.(((((....-..))))).))).((..((-(..((((((((---------.(((...(((......)))..)))))))).)))..)))...)).. ( -27.10, z-score = -1.47, R) >droWil1.scaffold_181136 1841328 106 - 2313701 CCUACUCAAAGAAUAAUUCGUU-UCACUUCUUGCACAAGUGUGAAUGCUGGUGAGAGGUCCC-----------UUGCUAUUUUUACCAUUUCCAUUGAUGGCCACUCGUGCGCAGCUC ..........(((........)-)).......((((.((((((((...(((..((......)-----------)..)))..))))(((((......))))).)))).))))....... ( -21.40, z-score = 0.44, R) >droVir3.scaffold_13049 7451190 113 - 25233164 CCUACUCCAGAGACAAUGGUGGUACCCUCUUCUGUCGAGUGUGAGUACCCGC-AGAGCUAUAGC----AAAAGCAAAUCUUGGCAUUUAUUUUAUUGACGAGCACUCGUGCAUAGCUC ..(((((((..((((..((.((...)).))..))))...)).))))).....-.(((((((.((----....((........)).............((((....))))))))))))) ( -28.40, z-score = -0.45, R) >droMoj3.scaffold_6680 1921106 111 - 24764193 CCUAUUCUAUAGACAAUGCC--UGCGCUCUUCAGUCGAGUGUGAGUACCAGC-AGAGGUCCGGC----AAAAGUAAAUCUUGGAAUAUCGUUUAUUGAUAGGCAGUGGUGCAUAGCUC ((((((..((((((..((((--.((((((.......))))))..(.(((...-...))).))))----).(((.....)))........)))))).))))))................ ( -25.50, z-score = 0.70, R) >droGri2.scaffold_15110 6077302 113 - 24565398 CGUACUCCAUGAACAAUGGUGGUAGACUCUUCAGUCGAGUGUGAAUACCAGC-AGAGGUCUAGC----AAAAAUGAAUCUUGCCAUUUAACUUAUUGAUGGGCACUCGUGCAUAGCUC .((((.((((.....)))).....((((....))))((((((........((-((....)).))----..............(((((.........)))))))))))))))....... ( -25.50, z-score = 0.53, R) >consensus CCCUCUCAAAGGACAAUGCUUUCUCACUCGACCGUCGAGUGUGAAUGCCGGCGAGAGGUCCUGC____GAAGAAUGUUCGCAUUGUCCUGUCCAUUGAGAGGCACUCGUGCGUAGCUC .....((((.(((.........(.(((((.......))))).)...(((.......)))...............................))).))))...(((....)))....... ( -8.89 = -8.47 + -0.41)

| Location | 14,946,534 – 14,946,648 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.32 |
| Shannon entropy | 0.62989 |
| G+C content | 0.52886 |
| Mean single sequence MFE | -36.89 |
| Consensus MFE | -11.91 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
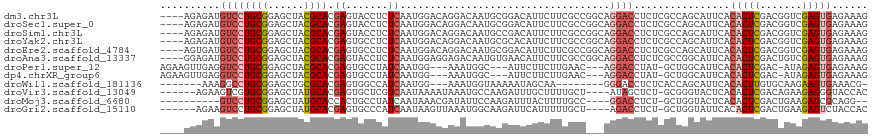
>dm3.chr3L 14946534 114 - 24543557 ----AGAGAUGUCCUGCGGAGCUACGCACGAGUACCUCUCAAUGGACAGGACAAUGCGGACAUUCUUCGCCGGCAGGACCUCUCGCCAGCAUUCACACUCGACGGUCGAGUGAGAAAG ----.((((.(((((((.(.((..((((...((.(((.((....)).)))))..))))((......))))).))))))).)))).......(((.((((((.....)))))).))).. ( -43.90, z-score = -2.53, R) >droSec1.super_0 7064937 114 - 21120651 ----AGAGAUGUCCUGCGGAGCUACGCACGAGUACCUCUCAAUGGACAGGACAAUGCGGACAUUCUUCGCCGGCAGGACCUCUCGCCAGCAUUCACACUCGACGGUCGAGUGAGAAAG ----.((((.(((((((.(.((..((((...((.(((.((....)).)))))..))))((......))))).))))))).)))).......(((.((((((.....)))))).))).. ( -43.90, z-score = -2.53, R) >droSim1.chr3L 14264840 114 - 22553184 ----AGAGAUGUCCUGCGGAGCUACGCACGAGUACCUCUCAAUGGACAGGACAAUGCCGACAUUCUUCGCCGGCAGGACCUCUCGCCAGCAUUCACACUCGACGGUCGAGUGAGAAAG ----.((((.(((((((((((.(((......))).))))...(((...(((.((((....)))).))).)))))))))).)))).......(((.((((((.....)))))).))).. ( -42.70, z-score = -2.39, R) >droYak2.chr3L 15035232 114 - 24197627 ----AGAGAUGUCCUGCGGAGCUACGCACGAGUGCCUCUCAAUGGACAGGACAAUGCGCACAUUCUUCGCCGGCAGGACCUCUCGCCAGCAUUCACACUCGACGGUCGAGUGAGAAAG ----.((((.(((((((((((.(((......))).))))...(((...(((.((((....)))).))).)))))))))).)))).......(((.((((((.....)))))).))).. ( -42.40, z-score = -1.77, R) >droEre2.scaffold_4784 14905624 114 - 25762168 ----AGUGAUGUCCUGCGGAGCUACGCACGAGUGCCUCUCAAUGGACAGGACAAUGCGGACAUUCUUCGCCGGCAGGACCUCUCGCCAGCAUUCACACUCGACGGUCGAGUGAGAAAG ----.((((.(((((((.(.((..((((....(((((.((....)).))).)).))))((......))))).)))))))...)))).....(((.((((((.....)))))).))).. ( -40.40, z-score = -0.88, R) >droAna3.scaffold_13337 19123918 114 + 23293914 ----GGAGAUGUCCUGCGGAGCUACGCACGAGUACCUCUCAAUGGAGGAGACAAUGUGAACAUUCUUCGCCGGCAGGACCUCUCGCCGGCAUUCACACUCGACUGUCGAGUGAGAAAG ----.((((.(((((((((((.(((......))).))))...(((.(((((..(((....)))))))).)))))))))).)))).......(((.((((((.....)))))).))).. ( -46.90, z-score = -3.18, R) >droPer1.super_12 943266 107 + 2414086 AGAAGUUGAGGUCCUGCGGAGCUACGCACGAGUGCCUAUCAAUGG---AAAUGGC---AUUCUUCUUGAAC---AGGACCUAU-GCUGGCAUUCACACUCGAC-AUAGAGUGAGAAAG .(.(((..((((((((((......)))..(((((((..((....)---)...)))---)))).........---)))))))..-)))..).(((.(((((...-...))))).))).. ( -38.10, z-score = -3.08, R) >dp4.chrXR_group6 12526137 107 + 13314419 AGAAGUUGAGGUCCUGCGGAGCUACGCACGAGUGCCUAUCAAUGG---AAAUGGC---AUUCUUCUUGAAC---AGGACCUAU-GCUGGCAUUCACACUCGAC-AUAGAGUGAGAAAG .(.(((..((((((((((......)))..(((((((..((....)---)...)))---)))).........---)))))))..-)))..).(((.(((((...-...))))).))).. ( -38.10, z-score = -3.08, R) >droWil1.scaffold_181136 1841346 99 + 2313701 -------AAAGGCCUGCGGAGCUGCGCACGAGUGGCCAUCAAUGG---AAAUGGUAAAAAUAGCAA--------GGGACCUCUCACCAGCAUUCACACUUGUGCAAGAAGUGAAACG- -------........(((....)))((((((((((((((......---..))))).......((..--------((((....)).)).)).....))))))))).............- ( -24.90, z-score = 0.30, R) >droVir3.scaffold_13049 7451208 107 + 25233164 ------AGAAGUCGUGCGGAGCUAUGCACGAGUGCUCGUCAAUAAAAUAAAUGCCAAGAUUUGCUUUUGCU----AUAGCUCU-GCGGGUACUCACACUCGACAGAAGAGGGUACCAC ------........((((((((((((((.(((((...(((.................))).))))).))).----))))))))-)))(((((((.(..((....)).).))))))).. ( -32.93, z-score = -1.64, R) >droMoj3.scaffold_6680 1921124 101 + 24764193 ----------GUCCUGCGGAGCUAUGCACCACUGCCUAUCAAUAAACGAUAUUCCAAGAUUUACUUUUGCC----GGACCUCU-GCUGGUACUCACACUCGACUGAAGAGCGCAGG-- ----------..(((((((((....(((....))).((((.......))))................((((----((......-.)))))))))...(((.......)))))))))-- ( -21.50, z-score = -0.05, R) >droGri2.scaffold_15110 6077320 107 + 24565398 ------AGAAGUCCUGCGGAGCUAUGCACGAGUGCCCAUCAAUAAGUUAAAUGGCAAGAUUCAUUUUUGCU----AGACCUCU-GCUGGUAUUCACACUCGACUGAAGAGUCUACCAC ------.(((..((.((((((....(((....)))................((((((((.....)))))))----)...))))-)).))..)))......((((....))))...... ( -26.90, z-score = -0.70, R) >consensus ____AGAGAUGUCCUGCGGAGCUACGCACGAGUGCCUCUCAAUGGACAAGACAAUGCGAACAUUCUUCGCC___AGGACCUCUCGCCAGCAUUCACACUCGACGGUCGAGUGAGAAAG ..........((((((((......)))).((.......))...................................))))................(((((.......)))))...... (-11.91 = -12.32 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:18 2011