| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,254,133 – 5,254,231 |
| Length | 98 |
| Max. P | 0.943006 |

| Location | 5,254,133 – 5,254,231 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Shannon entropy | 0.46873 |
| G+C content | 0.53353 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -18.79 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
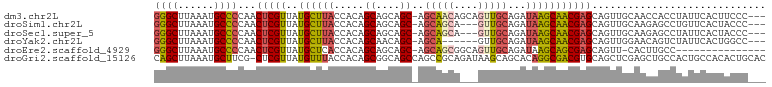
>dm3.chr2L 5254133 98 + 23011544 GGGCUUAAAUGCCCCAACUCGUUAUGCUUACCACAGCAGCAGC-AGCAACAGCAGUUGCAGAUAAGCAACGAGCAGUUGCAACCACCUAUUCACUUCCC--- ((((......))))......(((.((((......))))..)))-.(((((.((.(((((......)))))..)).)))))...................--- ( -29.20, z-score = -3.00, R) >droSim1.chr2L 5052631 95 + 22036055 GGGCUUAAAUGCCCCAACUCGUUAUGCUUACCACAGCAGCAGC-AGCAGCA---GUUGCAGAUAAGCAACGAGCAGUUGCAAGAGCCUGUUCACUACCC--- ((((......))))...(((((..((((......)))))).((-(((.((.---(((((......)))))..)).)))))..)))..............--- ( -29.60, z-score = -1.75, R) >droSec1.super_5 3334328 95 + 5866729 GGGCUUAAAUGCCCCAACUCGUUAUGCUUACCACAGCAGCAGC-AGCAGCA---GUUGCAGAUAAGCAACGAGCAGUUGCAAGAGCCUAUUCACUACCC--- ((((......))))...(((((..((((......)))))).((-(((.((.---(((((......)))))..)).)))))..)))..............--- ( -29.60, z-score = -2.39, R) >droYak2.chr2L 8379798 92 - 22324452 GGGCUUAAAUGCCCCAACUCGUUAUGCUUACCACAGCAACAGC-AGCA------GUUGCAGAUAAGCAACGAGCAGUUGGAACAGUCUAUUCACUGGCC--- ((((......))))(((((.(((.((((((.(...(((((...-....------))))).).))))))...))))))))...((((......))))...--- ( -27.80, z-score = -1.72, R) >droEre2.scaffold_4929 5332572 85 + 26641161 GGGCUUAAAUGCCCCAACUCGUUAUGCUCACCACAGCAGCAGC-AGCAGCGGCAGUUGCAGAUAAGCAGCGAGCAGUU-CACUUGCC--------------- ((((......))))......(...(((((.........((.((-....)).)).(((((......))))))))))...-).......--------------- ( -26.70, z-score = -0.59, R) >droGri2.scaffold_15126 999364 101 + 8399593 CAGCUUAAAUGCUUCG-CUCGUUAUGUUUACCACAGCGGCAGCCAGCCGCAGAUAAGCAGCACAGGCGACGUGCAGCUCGAGCUGCCACUGCCACACUGCAC ..((......((.(((-(..((..((((((.(...(((((.....))))).).))))))..))..)))).(((((((....)))).))).))......)).. ( -33.60, z-score = -0.43, R) >consensus GGGCUUAAAUGCCCCAACUCGUUAUGCUUACCACAGCAGCAGC_AGCAGCA___GUUGCAGAUAAGCAACGAGCAGUUGCAACAGCCUAUUCACUACCC___ ((((......))))...(((((..((((((.....((....))..(((((....)))))...)))))))))))............................. (-18.79 = -19.27 + 0.48)
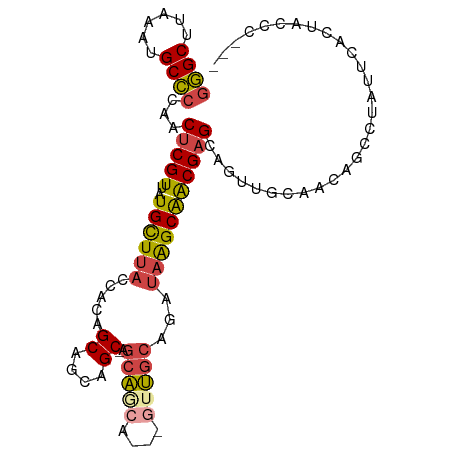
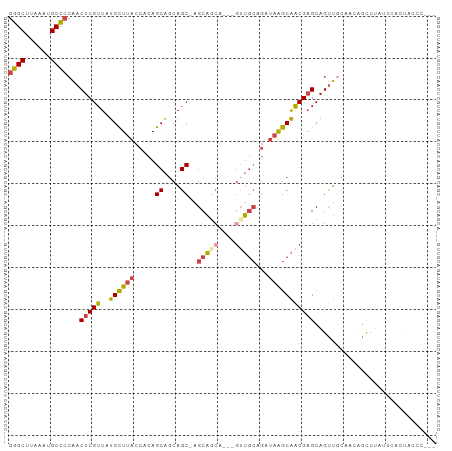
| Location | 5,254,133 – 5,254,231 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Shannon entropy | 0.46873 |
| G+C content | 0.53353 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -19.51 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5254133 98 - 23011544 ---GGGAAGUGAAUAGGUGGUUGCAACUGCUCGUUGCUUAUCUGCAACUGCUGUUGCU-GCUGCUGCUGUGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCC ---............(((...(((..(.((((((((((((((.(((((....))))).-((....))...))))))))..)))))).)..))).....))). ( -34.20, z-score = -1.11, R) >droSim1.chr2L 5052631 95 - 22036055 ---GGGUAGUGAACAGGCUCUUGCAACUGCUCGUUGCUUAUCUGCAAC---UGCUGCU-GCUGCUGCUGUGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCC ---............((((..(((..(.((((((((((((((.(((.(---.((....-)).).)))...))))))))..)))))).)..)))....)))). ( -35.10, z-score = -1.24, R) >droSec1.super_5 3334328 95 - 5866729 ---GGGUAGUGAAUAGGCUCUUGCAACUGCUCGUUGCUUAUCUGCAAC---UGCUGCU-GCUGCUGCUGUGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCC ---............((((..(((..(.((((((((((((((.(((.(---.((....-)).).)))...))))))))..)))))).)..)))....)))). ( -35.10, z-score = -1.48, R) >droYak2.chr2L 8379798 92 + 22324452 ---GGCCAGUGAAUAGACUGUUCCAACUGCUCGUUGCUUAUCUGCAAC------UGCU-GCUGUUGCUGUGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCC ---(((.(((......)))((((((...((((((((((((((.(((((------....-...)))))...))))))))..))))))))))))......))). ( -35.30, z-score = -2.84, R) >droEre2.scaffold_4929 5332572 85 - 26641161 ---------------GGCAAGUG-AACUGCUCGCUGCUUAUCUGCAACUGCCGCUGCU-GCUGCUGCUGUGGUGAGCAUAACGAGUUGGGGCAUUUAAGCCC ---------------((((((((-..(((((((.((((((((.(((.(.((.((....-)).)).).)))))))))))...)))).)))..)))))..))). ( -31.40, z-score = -0.76, R) >droGri2.scaffold_15126 999364 101 - 8399593 GUGCAGUGUGGCAGUGGCAGCUCGAGCUGCACGUCGCCUGUGCUGCUUAUCUGCGGCUGGCUGCCGCUGUGGUAAACAUAACGAG-CGAAGCAUUUAAGCUG (..(((.(((((.(((.((((....)))))))))))))))..).(((((..((((((.....)))((((((.....)))....))-)...)))..))))).. ( -42.50, z-score = -0.64, R) >consensus ___GGGUAGUGAAUAGGCUGUUGCAACUGCUCGUUGCUUAUCUGCAAC___UGCUGCU_GCUGCUGCUGUGGUAAGCAUAACGAGUUGGGGCAUUUAAGCCC ..........................(.((((((((((((((.(((........)))..((....))...))))))))..)))))).)((((......)))) (-19.51 = -20.35 + 0.84)

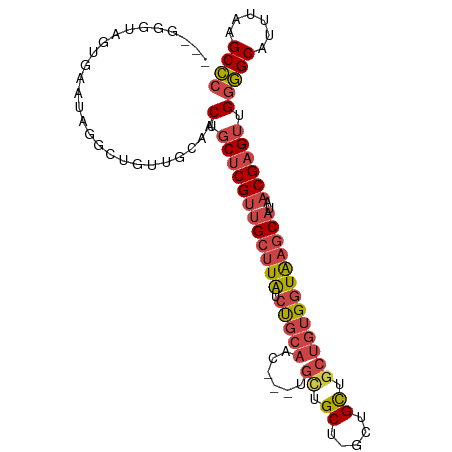
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:32 2011