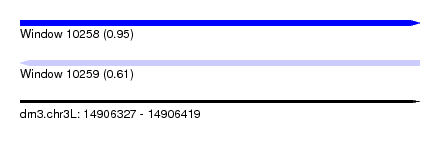
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,906,327 – 14,906,419 |
| Length | 92 |
| Max. P | 0.949488 |

| Location | 14,906,327 – 14,906,419 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Shannon entropy | 0.51165 |
| G+C content | 0.46740 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -13.36 |
| Energy contribution | -12.04 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
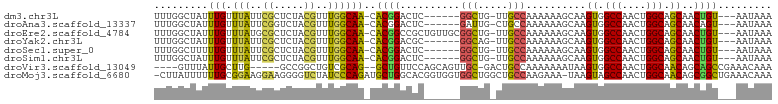
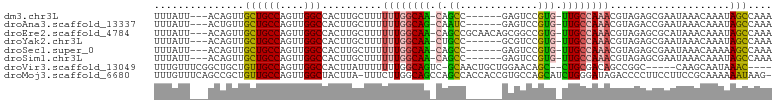
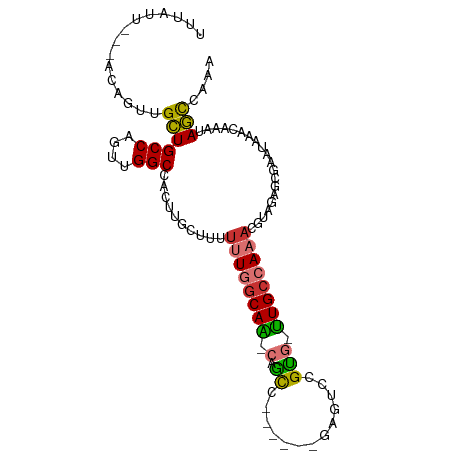
>dm3.chr3L 14906327 92 + 24543557 UUUGGCUAUUUGUUUAUUCGCUCUACGUUUGGCAA-CACGGACUC------GGCUG-UUGCCAAAAAAGCAAGUGGCCAACUGGCAGCAACUGU---AAUAAA .((((((((((((((...((.....))((((((((-((((....)------)..))-)))))))).))))))))))))))...((((...))))---...... ( -31.20, z-score = -2.80, R) >droAna3.scaffold_13337 19085239 92 - 23293914 UUUGGCUAUUUGUUUAUUCGGUCUACGUUUGGCAA-CACGGACUC------GAUUG-CUGCCAAAAAAGCAAGUGGCCAACUGGCAGCAACAGU---AAUAAA ..(((((((((((((...((.....))(((((((.-((((....)------)..))-.))))))).)))))))))))))((((.......))))---...... ( -29.50, z-score = -2.60, R) >droEre2.scaffold_4784 14866430 98 + 25762168 UUUGGCUAUUUGUUUAUGCGCUCUACGUUUGGCAA-CACGGCCGCUGUUGCGGCUG-UUGCCAAAAAAGCAAGUGGCCAACUGGCAGCAACUGU---AAUAAA .((((((((((((((..(((.....)))(((((((-((..(((((....)))))))-)))))))..))))))))))))))...((((...))))---...... ( -42.80, z-score = -3.94, R) >droYak2.chr3L 14992032 92 + 24197627 UUUGGCUAUUUGUUUAUUCGCUCUACGUUUGGCAA-CACGGACGC------GGCAG-UUGCCAAAAAAGCAAGUGGCCAACUGGCAGCAACUGU---AAUAAA .((((((((((((((...((.....))((((((((-(.((....)------)...)-)))))))).))))))))))))))...((((...))))---...... ( -31.00, z-score = -2.18, R) >droSec1.super_0 7024485 92 + 21120651 UUUGGCUUUUUGUUUAUUCGCUCUACGUUUGGCAA-CACGGACUC------GGCUG-UUGCCAAAAAAGCAAGUGGCCAACUGGCAGCAACUGU---AAUAAA .((((((.(((((((...((.....))((((((((-((((....)------)..))-)))))))).))))))).))))))...((((...))))---...... ( -27.10, z-score = -1.30, R) >droSim1.chr3L 14222514 92 + 22553184 UUUGGCUAUUUGUUUAUUCGCUCUACGUUUGGCAA-CACGGACUC------GGCUG-UUGCCAAAAAAGCAAGUGGCCAACUGGCAGCAACUGU---AAUAAA .((((((((((((((...((.....))((((((((-((((....)------)..))-)))))))).))))))))))))))...((((...))))---...... ( -31.20, z-score = -2.80, R) >droVir3.scaffold_13049 7418424 91 - 25233164 ----GUUUAUUGCUUG-----GCCGGCUGUCGCAG--GCUGUUCCAGCAGUUGC-GACUGCCAAAAAAAUAAGUGGCCAACUGGCAACAGCAGCCGAAACAAA ----((((...(((((-----((((((.(((((((--.((((....))))))))-))).)))............))))).(((....))).)))..))))... ( -36.30, z-score = -2.49, R) >droMoj3.scaffold_6680 1857076 101 - 24764193 -CUUAUUUUUUGCGGAAGGAAGGGGUCUAUCCCAGAUGCUGGCACGGUGGUGGCUGGCUGCCAAGAAA-UAAGUAGCCAACUGGCAACAGCGGCUGAAACAAA -(((((((((((((.......((((....))))....((..((((....)).))..)))).)))))))-))))(((((..(((....))).)))))....... ( -34.40, z-score = -1.15, R) >consensus UUUGGCUAUUUGUUUAUUCGCUCUACGUUUGGCAA_CACGGACUC______GGCUG_UUGCCAAAAAAGCAAGUGGCCAACUGGCAGCAACUGU___AAUAAA .........(((.(((..((.....))..))))))..((((..........(((.....)))..........((.(((....))).))..))))......... (-13.36 = -12.04 + -1.33)
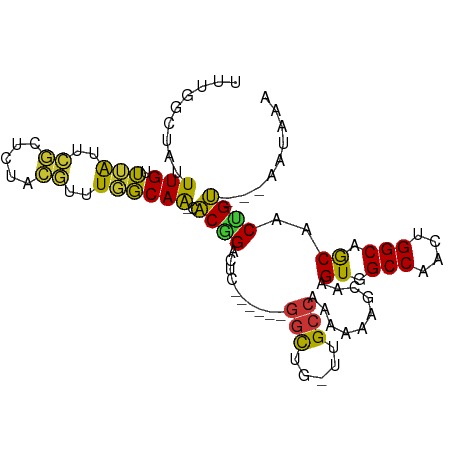
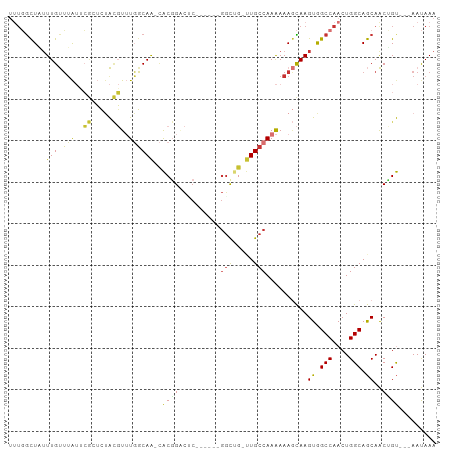
| Location | 14,906,327 – 14,906,419 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 74.51 |
| Shannon entropy | 0.51165 |
| G+C content | 0.46740 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -11.94 |
| Energy contribution | -11.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14906327 92 - 24543557 UUUAUU---ACAGUUGCUGCCAGUUGGCCACUUGCUUUUUUGGCAA-CAGCC------GAGUCCGUG-UUGCCAAACGUAGAGCGAAUAAACAAAUAGCCAAA ......---......((((((....)))...(((((((((((((((-((..(------(....))))-))))))))...)))))))..........))).... ( -25.00, z-score = -1.29, R) >droAna3.scaffold_13337 19085239 92 + 23293914 UUUAUU---ACUGUUGCUGCCAGUUGGCCACUUGCUUUUUUGGCAG-CAAUC------GAGUCCGUG-UUGCCAAACGUAGACCGAAUAAACAAAUAGCCAAA ......---.(((((((((((((..(((.....)))...)))))))-)..((------(.(((..((-((....))))..)))))).......)))))..... ( -29.60, z-score = -3.30, R) >droEre2.scaffold_4784 14866430 98 - 25762168 UUUAUU---ACAGUUGCUGCCAGUUGGCCACUUGCUUUUUUGGCAA-CAGCCGCAACAGCGGCCGUG-UUGCCAAACGUAGAGCGCAUAAACAAAUAGCCAAA ......---.....(((.(((....))).....(((((((((((((-(((((((....)))))..))-))))))))...))))))))................ ( -34.30, z-score = -2.20, R) >droYak2.chr3L 14992032 92 - 24197627 UUUAUU---ACAGUUGCUGCCAGUUGGCCACUUGCUUUUUUGGCAA-CUGCC------GCGUCCGUG-UUGCCAAACGUAGAGCGAAUAAACAAAUAGCCAAA ......---......((((((....)))...(((((((((((((((-(...(------(....)).)-))))))))...)))))))..........))).... ( -23.40, z-score = -0.27, R) >droSec1.super_0 7024485 92 - 21120651 UUUAUU---ACAGUUGCUGCCAGUUGGCCACUUGCUUUUUUGGCAA-CAGCC------GAGUCCGUG-UUGCCAAACGUAGAGCGAAUAAACAAAAAGCCAAA ......---......((((((....)))...(((((((((((((((-((..(------(....))))-))))))))...)))))))..........))).... ( -25.10, z-score = -1.22, R) >droSim1.chr3L 14222514 92 - 22553184 UUUAUU---ACAGUUGCUGCCAGUUGGCCACUUGCUUUUUUGGCAA-CAGCC------GAGUCCGUG-UUGCCAAACGUAGAGCGAAUAAACAAAUAGCCAAA ......---......((((((....)))...(((((((((((((((-((..(------(....))))-))))))))...)))))))..........))).... ( -25.00, z-score = -1.29, R) >droVir3.scaffold_13049 7418424 91 + 25233164 UUUGUUUCGGCUGCUGUUGCCAGUUGGCCACUUAUUUUUUUGGCAGUC-GCAACUGCUGGAACAGC--CUGCGACAGCCGGC-----CAAGCAAUAAAC---- .((((((.((((((((((((..((((...........(((..(((((.-...)))))..)))))))--..))))))).))))-----))))))).....---- ( -36.50, z-score = -2.81, R) >droMoj3.scaffold_6680 1857076 101 + 24764193 UUUGUUUCAGCCGCUGUUGCCAGUUGGCUACUUA-UUUCUUGGCAGCCAGCCACCACCGUGCCAGCAUCUGGGAUAGACCCCUUCCUUCCGCAAAAAAUAAG- (((((..(((..((((......(((((((.((..-......)).)))))))(((....))).))))..)))..)))))........................- ( -25.90, z-score = -0.83, R) >consensus UUUAUU___ACAGUUGCUGCCAGUUGGCCACUUGCUUUUUUGGCAA_CAGCC______GAGUCCGUG_UUGCCAAACGUAGAGCGAAUAAACAAAUAGCCAAA ...............((((((....)))..........((((((((...((.............))..))))))))....................))).... (-11.94 = -11.38 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:08 2011