| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,882,289 – 14,882,400 |
| Length | 111 |
| Max. P | 0.886255 |

| Location | 14,882,289 – 14,882,400 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 81.18 |
| Shannon entropy | 0.36530 |
| G+C content | 0.36382 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -14.64 |
| Energy contribution | -15.30 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
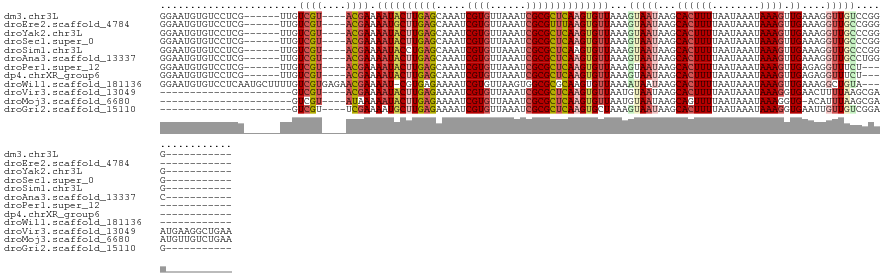
>dm3.chr3L 14882289 111 - 24543557 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGUCCGGG----------- (((..........(------((....(----((...(((((((((((.....(((......))))))))))))))...)))...))).((((((((.......))))))))....)))...----------- ( -27.80, z-score = -2.81, R) >droEre2.scaffold_4784 14842452 110 - 25762168 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUGCUUGAGCAAAUCGUGUUAAAUCGCGUUUAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGCCGGG------------ (((.....)))(((------(......----))))...(((((.((((...((((......))))......)))).)))))....(((((((((((.......))))))))..)))....------------ ( -20.90, z-score = -0.21, R) >droYak2.chr3L 14967101 111 - 24197627 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGCCCGGG----------- ((...........(------((....(----((...(((((((((((.....(((......))))))))))))))...)))...))).((((((((.......)))))))).....))...----------- ( -26.30, z-score = -1.95, R) >droSec1.super_0 6995021 111 - 21120651 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGCCCGGG----------- ((...........(------((....(----((...(((((((((((.....(((......))))))))))))))...)))...))).((((((((.......)))))))).....))...----------- ( -26.30, z-score = -1.95, R) >droSim1.chr3L 14198321 111 - 22553184 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUACCUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGCCCGGG----------- (((.....)))(((------(......----)))).....((((.((((....(((((....((((....)))).....)))))....((((((((.......)))))))).)))).))))----------- ( -23.00, z-score = -0.83, R) >droAna3.scaffold_13337 19060757 111 + 23293914 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGCCUGGC----------- (((.....)))..(------((....(----((...(((((((((((.....(((......))))))))))))))...)))...))).((((((((.......))))))))..........----------- ( -26.10, z-score = -1.93, R) >droPer1.super_12 877141 107 + 2414086 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAGAGGUUUCU--------------- ((((.....(((((------((....(----((...(((((((((((.....(((......))))))))))))))...)))...)))(((((........)))))...)))).))))--------------- ( -27.50, z-score = -3.09, R) >dp4.chrXR_group6 12457613 107 + 13314419 GGAAUGUGUCCUCG------UUGUCGU----ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAGAGGUUUCU--------------- ((((.....(((((------((....(----((...(((((((((((.....(((......))))))))))))))...)))...)))(((((........)))))...)))).))))--------------- ( -27.50, z-score = -3.09, R) >droWil1.scaffold_181136 1582764 116 + 2313701 GGAAUGUGUCCUCAAUGCUUUUGUCGUGAGAACGAAAAU-CGUGAGAAAAUCGUGUUAAGUGGCGCGCAAGUGUUAAAAUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGCUGUA--------------- ..........((((....(((((((....).))))))..-..)))).......(((((..((((((....))))))...)))))(((.((((((((.......)))))))))))...--------------- ( -28.90, z-score = -2.54, R) >droVir3.scaffold_13049 7399140 106 + 25233164 ----------------------GUCGU----ACGAAAAUACUUGAGAAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAUGUAAUAAGCACUUUUAAUAAAUAAAGGUGAACUUUUAAGCGAAUGAAGGCUGAA ----------------------....(----(((..((((((((((.....((((......))))))))))))))..))))...(((.((((.......((((((....)))))).......)))))))... ( -25.94, z-score = -2.73, R) >droMoj3.scaffold_6680 1833981 105 + 24764193 ----------------------GUCGU----AUAAAAAUACUUGAGAAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAUGUAAUAAGCAGUUUUAAUAAAUAAAGGUG-ACAUUUAAGCGAAUGUUGUCUGAA ----------------------((..(----(((..((((((((((.....((((......))))))))))))))..))))....))...............(((..-((((((....))))))..)))... ( -24.20, z-score = -2.93, R) >droGri2.scaffold_15110 6027074 95 + 24565398 ----------------------GUCGU----UCGAAAAUGCUUGAGAAAAUCGUGUUAAAUCGCGCUCAAGUGCUAAAGUAAUAAGCACUUUUAAUAAAUAAAGGUGAAUUGUUGUCGGAG----------- ----------------------....(----((((....(((((((.....((((......))))))))))).......((((((.((((((((.....))))))))..))))))))))).----------- ( -22.70, z-score = -2.33, R) >consensus GGAAUGUGUCCUCG______UUGUCGU____ACGAAAAUACUUGAGCAAAUCGUGUUAAAUCGCGCUCAAGUGUUAAAGUAAUAAGCACUUUUAAUAAAUAAAGUUGAAAGGUUGCCCGGG___________ .......................((((....)))).((((((((((.....((((......))))))))))))))...(((((...((((((........)))).))....)))))................ (-14.64 = -15.30 + 0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:07 2011