
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,875,714 – 14,875,840 |
| Length | 126 |
| Max. P | 0.864673 |

| Location | 14,875,714 – 14,875,804 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.93 |
| Shannon entropy | 0.57096 |
| G+C content | 0.55577 |
| Mean single sequence MFE | -17.46 |
| Consensus MFE | -7.92 |
| Energy contribution | -8.45 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
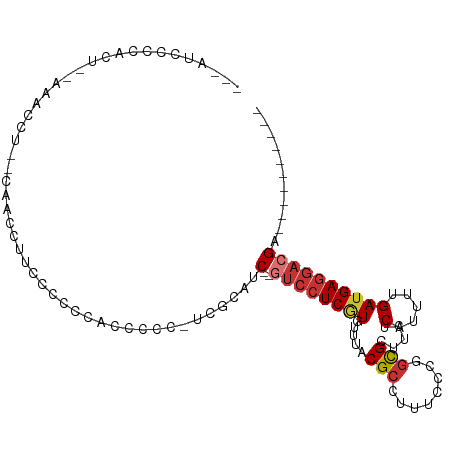
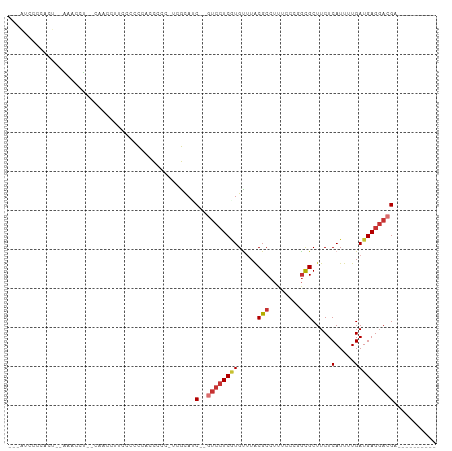
>dm3.chr3L 14875714 90 + 24543557 ---AAACCCACU--GAACCU--CAACCUUCGCCCCCCCCCC-UCGCAAC--GUCCUCGUGUUUACGCCUUUCCCUGCGCUUCUCAUUUUGAUGAGGACGA---------- ---.........--......--...................-......(--((((((((.....(((........))).....(.....)))))))))).---------- ( -14.50, z-score = -1.63, R) >droPer1.super_12 870456 95 - 2414086 --------CAGUACACCCCUG-CCAGCCUCCCGCAACCGAC-UAGCAUC--AUCCUCAUGUUUACGCCUUUCUCGGUGCUUCUCAUUUUGAUGAGGAGGACGAUGCC--- --------.............-...((.....)).......-..(((((--.(((((.......((((......))))...(((((....)))))))))).))))).--- ( -24.90, z-score = -1.60, R) >dp4.chrXR_group6 12450952 98 - 13314419 --------CAGUACACCCCUG-CCAGCCUCCCGCAACCGAC-UAGCAUC--AUCCUCAUGUUUACGCCUUUCUCGGUGCUUCUCAUUUUGAUGAGGAGGACGAUGCCAGA --------..........(((-...((.....)).......-..(((((--.(((((.......((((......))))...(((((....)))))))))).)))))))). ( -26.60, z-score = -1.62, R) >droAna3.scaffold_13337 19054492 100 - 23293914 CACGCCGCUGCCUUGAUCCUGUCUCCAUUCCCCUCGCUGCCAUUACAACCAGCCAUCCUGUAUGCGCCAUCCUCGACGCUUCUCAUUUUGAUGAGGACGA---------- ...(.((((((...(((...)))............((((..........))))......))).))).)........((.(((((((....))))))))).---------- ( -14.90, z-score = 1.01, R) >droEre2.scaffold_4784 14835896 87 + 25762168 ---AUCCCCACU--GAACCC--CACCUUCGUCCC--UCCCC--CGCACC--GUCCUCGUGCUUACGCCUUUCCCAGCGCUUCUCAUUUUGAUGAGCACGA---------- ---.........--......--............--.....--......--....((((((((((((........)))....((.....)))))))))))---------- ( -13.10, z-score = -1.88, R) >droYak2.chr3L 14960157 88 + 24197627 ---AUCCCCACU--GAAUCU--CCCCACUUUCUC--CCCCC-UCGCAUC--GUCCUCGUGCUUACGCCUUUACCAGCGCUUCUCAUUUUGAUGAGGACGA---------- ---.........--......--............--.....-.....((--(((((((((((............)))))...((.....)).))))))))---------- ( -17.20, z-score = -2.83, R) >droSec1.super_0 6988369 92 + 21120651 ---AUCCCUACUGAAAACCU--UAACCUUCCCCCCACCCCC-UCGUACC--GUCCUCGUGUUUACGCCUUUCCCUGCGCUUCUCAUUUUGAUGAGGACGA---------- ---.................--...................-......(--((((((((.....(((........))).....(.....)))))))))).---------- ( -14.30, z-score = -2.43, R) >droSim1.chr3L 14191669 90 + 22553184 ---AUCCCCACU--AAACCU--CAACCUUCCCCCCACCCCC-UCGUAGC--GUCCUCGUGUUUACGCCUUUCCCUGCGCUUCUCAUUUUGAUGAGGACGA---------- ---.........--......--...................-......(--((((((((.....(((........))).....(.....)))))))))).---------- ( -14.20, z-score = -1.98, R) >consensus ___AUCCCCACU__AAACCU__CAACCUUCCCCCCACCCCC_UCGCAUC__GUCCUCGUGUUUACGCCUUUCCCGGCGCUUCUCAUUUUGAUGAGGACGA__________ ...................................................((((((((.....(((........))).....(.....)))))))))............ ( -7.92 = -8.45 + 0.53)

| Location | 14,875,746 – 14,875,840 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.55 |
| Shannon entropy | 0.51504 |
| G+C content | 0.53527 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -14.11 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
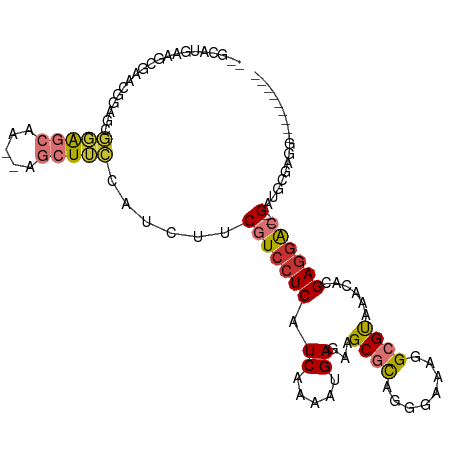
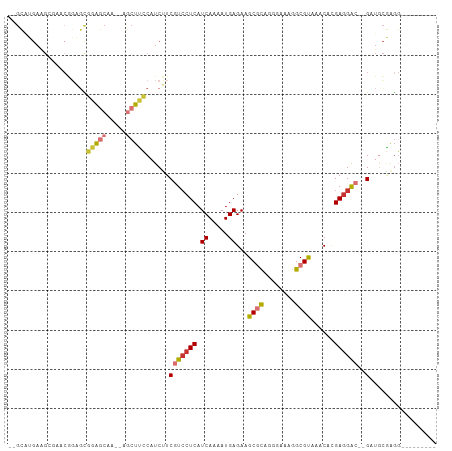
>dm3.chr3L 14875746 94 - 24543557 --GCAUGAAGCGAACGGAGCGGAGCAA--AGCUUCCAUCUUCGUCCUCAUCAAAAUGAGAAGCGCAGGGAAAGGCGUAAACACGAGGAC--GUUGCGAGG--------- --(((.((((.....(((((.......--.)))))...))))((((((.((.....))...((((........))))......))))))--..)))....--------- ( -27.20, z-score = -0.56, R) >droPer1.super_12 870486 91 + 2414086 GAGAGGGAGG-------CUUCCAUCUUCCAUCUGGCAUCGUCCUCCUCAUCAAAAUGAGAAGCACCGAGAAAGGCGUAAACAUGAGGAU--GAUGCUAGU--------- ....((((((-------......))))))..(((((((((((((((((((....)))))..((.((......)).))......))))))--)))))))).--------- ( -37.60, z-score = -4.03, R) >dp4.chrXR_group6 12450982 98 + 13314419 GAGAGGGAGGCUUCCAUCUUCCAUCUUCCAUCUGGCAUCGUCCUCCUCAUCAAAAUGAGAAGCACCGAGAAAGGCGUAAACAUGAGGAU--GAUGCUAGU--------- ((((((((((......)))))).))))....(((((((((((((((((((....)))))..((.((......)).))......))))))--)))))))).--------- ( -39.40, z-score = -3.85, R) >droAna3.scaffold_13337 19054523 94 + 23293914 -------------ACAGAGACCGGCAU--AGCUUCCAUCUUCGUCCUCAUCAAAAUGAGAAGCGUCGAGGAUGGCGCAUACAGGAUGGCUGGUUGUAAUGGCAGCGAGG -------------.((..(((((((..--.((..(((((((((.(((..((.....))..)).).))))))))).))...(.....))))))))....))......... ( -29.80, z-score = -0.87, R) >droEre2.scaffold_4784 14835925 94 - 25762168 --GCCUGAAGAGUACGGAGCGGAGCAA--AGCUUCCAUCUUCGUGCUCAUCAAAAUGAGAAGCGCUGGGAAAGGCGUAAGCACGAGGAC--GGUGCGGGG--------- --.(((((.((((((((((.(((((..--.)))))...)))))))))).))..........((((((.....(((....)).).....)--)))))))).--------- ( -37.10, z-score = -2.72, R) >droYak2.chr3L 14960187 94 - 24197627 --GCAUGAAGCGAACGGUGCGGAGCGG--AGCUUCCAUCUUCGUCCUCAUCAAAAUGAGAAGCGCUGGUAAAGGCGUAAGCACGAGGAC--GAUGCGAGG--------- --((((.....(((.((((.(((((..--.))))))))))))((((((.((.....))...(((((......)))))......))))))--.))))....--------- ( -32.30, z-score = -1.14, R) >droSec1.super_0 6988403 94 - 21120651 --GCAUGAAGCGAACGGAGCGGAGCAA--AGCUUCCAUCUUCGUCCUCAUCAAAAUGAGAAGCGCAGGGAAAGGCGUAAACACGAGGAC--GGUACGAGG--------- --........((.(((((((.......--.)))))......(((((((.((.....))...((((........))))......))))))--))).))...--------- ( -25.30, z-score = -0.47, R) >droSim1.chr3L 14191701 94 - 22553184 --GCAUGAAGCGAACGGAGCGGAGCAA--AGCUUCCAUCUUCGUCCUCAUCAAAAUGAGAAGCGCAGGGAAAGGCGUAAACACGAGGAC--GCUACGAGG--------- --...((.((((...(((((.......--.)))))..(((((((.(((((....)))))..((((........))))....))))))))--))).))...--------- ( -27.80, z-score = -1.09, R) >consensus __GCAUGAAGCGAACGGAGCGGAGCAA__AGCUUCCAUCUUCGUCCUCAUCAAAAUGAGAAGCGCAGGGAAAGGCGUAAACACGAGGAC__GAUGCGAGG_________ ....................(((((.....))))).......((((((.((.....))...((((........))))......)))))).................... (-14.11 = -14.55 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:05 2011