| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,863,858 – 14,863,982 |
| Length | 124 |
| Max. P | 0.693831 |

| Location | 14,863,858 – 14,863,982 |
|---|---|
| Length | 124 |
| Sequences | 14 |
| Columns | 137 |
| Reading direction | reverse |
| Mean pairwise identity | 68.10 |
| Shannon entropy | 0.71179 |
| G+C content | 0.40874 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -12.09 |
| Energy contribution | -11.69 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.47 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
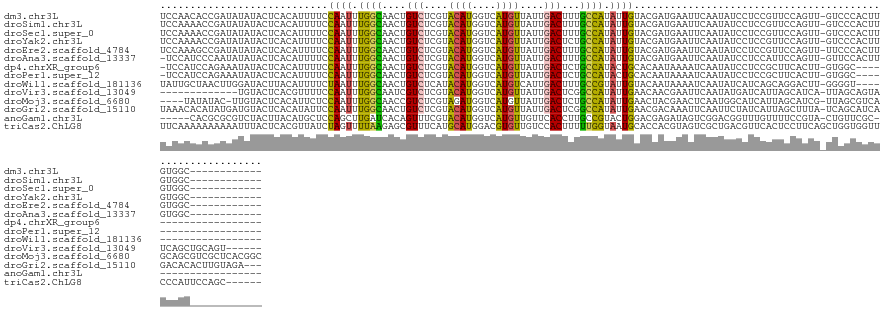
>dm3.chr3L 14863858 124 - 24543557 UCCAACACCGAUAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUUUGCCAUAUUGUACGAUGAAUUCAAUAUCCUCCGUUCCAGUU-GUCCCACUUGUGGC------------ ..((((...((.......)).((((...((((.((((((..(((....((((....))))....))).)))))).))))...)))).....................)))-)..(((....))).------------ ( -23.90, z-score = -0.66, R) >droSim1.chr3L 14179960 124 - 22553184 UCCAAAACCGAUAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUUUGCCAUAUUGUACGAUGAAUUCAAUAUCCUCCGUUCCAGUU-GUCCCACUUGUGGC------------ ..................................((((((((..(((((((((((...(((...)))...))))...)))))))((....)).............)))))-)))(((....))).------------ ( -23.50, z-score = -0.68, R) >droSec1.super_0 6976717 124 - 21120651 UCCAAAACCGAUAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUUUGCCAUAUUGUACGAUGAAUUCAAUAUCCUCCGUUCCAGUU-GUCCCACUUGUGGC------------ ..................................((((((((..(((((((((((...(((...)))...))))...)))))))((....)).............)))))-)))(((....))).------------ ( -23.50, z-score = -0.68, R) >droYak2.chr3L 14948304 124 - 24197627 UCCAAAACCGAUAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCUGCCAUAUUGUACGAUGAAUUCAAUAUCCUCCGUUCCAGUU-GUCCCACUUGUGGC------------ ..................................((((((((..(((((((((((...(((...)))...))))...)))))))((....)).............)))))-)))(((....))).------------ ( -23.50, z-score = -0.71, R) >droEre2.scaffold_4784 14824068 124 - 25762168 UCCAAAGCCGAUAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUUUGCCAUAUUGUACGAUGAAUUCAAUAUCCUCCGUUCCAGUU-UUCCCACUUGUGGC------------ ......((((((((.......((((...((((.((((((..(((....((((....))))....))).)))))).))))...))))......))))).........(((.-.....)))...)))------------ ( -24.02, z-score = -1.06, R) >droAna3.scaffold_13337 19043486 123 + 23293914 -UCCAUCCCAAUAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUUUGCCAUAUUGUACGAUGAAUUCAAUAUCCUCCAUUCCAGUU-GUUCCACUUGUGGC------------ -....................((((...((((.((((((..(((....((((....))))....))).)))))).))))...)))).............((((...(((.-.....))).)))).------------ ( -22.60, z-score = -0.82, R) >dp4.chrXR_group6 12440084 114 + 13314419 -UCCAUCCAGAAAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCUGCCAUACUGCACAAUAAAAUCAAUAUCCUCCGCUUCACUU-GUGGC--------------------- -................................(((((...(((....((((....))))....)))..))))).........................((((.......-)))).--------------------- ( -16.70, z-score = -0.08, R) >droPer1.super_12 859588 114 + 2414086 -UCCAUCCAGAAAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCUGCCAUACUGCACAAUAAAAUCAAUAUCCUCCGCUUCACUU-GUGGC--------------------- -................................(((((...(((....((((....))))....)))..))))).........................((((.......-)))).--------------------- ( -16.70, z-score = -0.08, R) >droWil1.scaffold_181136 1457132 115 + 2313701 UAUUGCUAACUUGGAUACUUACAUUUUCUAAUUUGGCAACUGUCUCAUACAUGGUCAUGUCAUUGACUUUGCCGUAUUGUACAAUAAAAUCAAUAUCAUCAGCAGGACUU-GGGGU--------------------- ..(((((((.(((((...........))))).)))))))...(((((.....((((.(((...(((..(((...((((....))))....)))..)))...))).)))))-)))).--------------------- ( -22.60, z-score = -0.58, R) >droVir3.scaffold_13049 7385009 117 + 25233164 -------------UGUACUCACGUUUUCCAAUUUGGCAAUCGUCUCGUACAUGGUCAUGUUAUUGACUCGGCCAUAUUGAACAACGAAUUCAAUAUGAUCAUUAGCAUCA-UUAGCAGUAUCAGCUGCAGU------ -------------(((((..(((.((.((.....)).)).)))...))))).(((((......))))).(..(((((((((.......)))))))))..)..........-...(((((....)))))...------ ( -28.70, z-score = -1.93, R) >droMoj3.scaffold_6680 1818588 131 + 24764193 ----UAUAUAC-UUGUACUCACAUUCUCCAAUUUGGCAACCGUCUCGUAGAUGGUCAUGUUAUUGACUCUGCCAUAUUGAACUACGAACUCAAUGGCAUCAUUAGCAUCG-UUAGCGUCAGCAGCGUCGCUCACGGC ----.......-.......................((.((((((.....)))))).((((((.(((...((((((..(((.........)))))))))))).))))))((-(.((((.(......).)))).))))) ( -29.20, z-score = -0.14, R) >droGri2.scaffold_15110 6014119 133 + 24565398 UAAACACAUAUGAUGUACUCACAUAUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCGGCCAUAUUGAACGACAAAUUCAAUUCUAUCAUUAGCUUUA-UCAGCAUCAGACACACUUGUAGA--- ..........((((((............((((.((((....(((....((((....))))....)))...)))).))))............((..(((....)))..)).-...))))))..............--- ( -22.10, z-score = 0.20, R) >anoGam1.chr3L 20454046 113 + 41284009 -----CACGCGCGUCUACUUACAUGCUCCAGCUUGAUCACAGUUUCGUACAUGGUCAUGUUGUUCACCUUGCCGUACUGGACGAGAUAGUCGGACGGUUUGUUUUCCGUA-CUGUUCGC------------------ -----...(((((((((..(((..((..((((.((((((..((.....)).)))))).))))........)).))).)))))).((((((((((..(....)..)))).)-))))))))------------------ ( -26.40, z-score = 1.10, R) >triCas2.ChLG8 13712819 131 + 15773733 UUCAAAAAAAAAAUUUACUCACGUUAUCUAGUUUUAAGAGCGUUUCAUGCAUGGACGUGUUGUCCACUUUUUGGUAAUGCACCACGUAGUCGCUGACGUUCACUCCUUCAGCUGGUGGUUCCCAUUCCAGC------ .....................................(((((((....((....(((((.((((((.....)))....))).)))))....)).))))))).........((((((((...)))..)))))------ ( -28.00, z-score = -0.44, R) >consensus UCCAAAACCGAUAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUUUGCCAUAUUGUACGAUGAAUUCAAUAUCCUCCGUUCCAGUU_GUCGCACUUGUGGC____________ ............................((((.((((....(((....((((....))))....)))...)))).)))).......................................................... (-12.09 = -11.69 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:27:04 2011