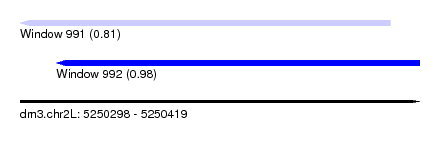
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,250,298 – 5,250,419 |
| Length | 121 |
| Max. P | 0.975517 |

| Location | 5,250,298 – 5,250,410 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Shannon entropy | 0.37435 |
| G+C content | 0.44262 |
| Mean single sequence MFE | -32.59 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.11 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5250298 112 - 23011544 UUUAUUUUGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCCAAUGGAGUGGGUACGGAUUUGG (((((((..(((..(..(..-((((((....))))))((((((.((.((((((........)))))).)).)))))).......)..)..)))..)))))))........... ( -30.80, z-score = -0.94, R) >droSim1.chr2L 5048820 112 - 22036055 UUUAUGUUGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCGAGUGGAGUGGGUACGGAUUUGG .............(((((((-((((((....))))))((((((.((.((((((........)))))).)).))))))((((.((........)).))))..)))))))..... ( -31.60, z-score = -0.87, R) >droSec1.super_5 3330633 112 - 5866729 UUUAUGUUGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCAGAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCGAGUGGAGUGGGUACGGAUUUGG .............(((((((-((((((....))))))((((((.((.((((((........)))))).)).))))))((((.((........)).))))..)))))))..... ( -31.60, z-score = -1.05, R) >droYak2.chr2L 8376192 111 + 22324452 -UUUGUGUGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAGAUGGAGUGGGCACGGAUUUGG -.((((((((((..(..(.(-((((((....))))))).)..)....))))).)))))((((((((.((((((((.(.(((.....))).).))))))))))))))))..... ( -36.40, z-score = -1.98, R) >droEre2.scaffold_4929 5328756 111 - 26641161 -UUUUUUUGUUAUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAUAUGGAGUGGGCACGGAUUUGG -......((((.........-..((((....))))((((.....))))))))(((.((((((((((.((((((((((.(((.....))).))))))))))))))))))))))) ( -33.80, z-score = -2.61, R) >droAna3.scaffold_12916 6266560 101 + 16180835 -----UUUGUUGUUUUGG---UUUUGGGGUACCGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGCCAUGGAGGAUUGGAGGUCGCAGAGGA---- -----(((((..(((..(---((((..((...((((.((((((.((.((((((........)))))).)).)))))).))))))....)))))..)))...)))))...---- ( -28.90, z-score = -0.68, R) >dp4.chr4_group3 7044101 101 - 11692001 ------UCUCUGUCUUAUUUCUUUUGGGGUAAGGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGGAUCCAGAGAGGAUUUGG------ ------.(((((((((((..(....)..)))).(((.((((((.((.((((((........)))))).)).)))))).))))))).)))(((((.....)))))...------ ( -33.80, z-score = -2.51, R) >droPer1.super_1 8496491 101 - 10282868 ------UCUCUGUCUUAUUUCUUUUGGGGUAAGGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGGAUCCAGAGAGGAUUUGG------ ------.(((((((((((..(....)..)))).(((.((((((.((.((((((........)))))).)).)))))).))))))).)))(((((.....)))))...------ ( -33.80, z-score = -2.51, R) >consensus _UU_UUUUGUUGUUUUGUAU_UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAAAUGGAGUGGGUACGGAUUUGG ..........(((((((.......))))))).((((.((((((.((.((((((........)))))).)).)))))).))))............................... (-21.77 = -21.11 + -0.65)

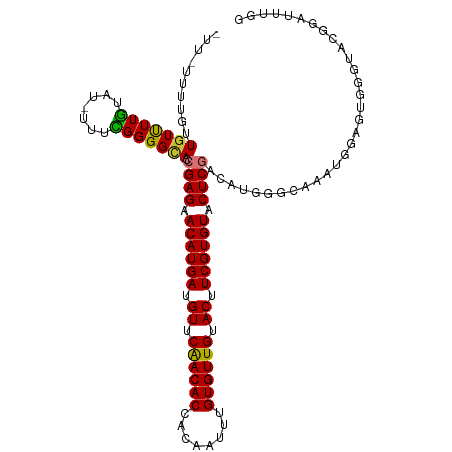
| Location | 5,250,309 – 5,250,419 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Shannon entropy | 0.37676 |
| G+C content | 0.42162 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.09 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
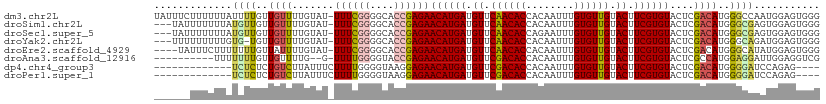
>dm3.chr2L 5250309 110 - 23011544 UAUUUCUUUUUUAUUUUGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCCAAUGGAGUGGG ..........((((((..(((..(..(..-((((((....))))))((((((.((.((((((........)))))).)).)))))).......)..)..)))..)))))). ( -30.70, z-score = -1.99, R) >droSim1.chr2L 5048831 107 - 22036055 ---UAUUUUUUUAUGUUGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCGAGUGGAGUGGG ---(((((.(((((((((.(((((((...-...))))))).)(((.((((((.((.((((((........)))))).)).)))))).))))))))))).)))))....... ( -31.30, z-score = -1.83, R) >droSec1.super_5 3330644 107 - 5866729 ---UAUUUUUUUAUGUUGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCAGAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCGAGUGGAGUGGG ---(((((.(((((((((.(((((((...-...))))))).)(((.((((((.((.((((((........)))))).)).)))))).))))))))))).)))))....... ( -31.30, z-score = -1.97, R) >droYak2.chr2L 8376203 106 + 22324452 ---UUUUUUUUUGUG-UGUUGUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAGAUGGAGUGGG ---.(((..((((((-(((((....((((-((((((....)))))))(((((.((.((((((........)))))).)).)))))))).))))))..)))))..))).... ( -31.00, z-score = -1.63, R) >droEre2.scaffold_4929 5328767 106 - 26641161 ----UAUUUCUUUUUUUGUUAUUUUGUAU-UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAUAUGGAGUGGG ----..............((((((..(((-.(((((....))(((.((((((.((.((((((........)))))).)).)))))).))).....))).)))..)))))). ( -29.30, z-score = -2.21, R) >droAna3.scaffold_12916 6266567 98 + 16180835 ----------UUUUUUUGUUGUUUUG--G-UUUUGGGGUACCGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGCCAUGGAGGAUUGGAGGUCG ----------.((((..(((.(((((--(-(...((....))(((.((((((.((.((((((........)))))).)).)))))).))))))).))).)))..))))... ( -27.80, z-score = -1.31, R) >dp4.chr4_group3 7044110 94 - 11692001 -------------UCUCUCUGUCUUAUUUCUUUUGGGGUAAGGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGGAUCCAGAG---- -------------(((((.((((((((..(....)..)))).(((.((((((.((.((((((........)))))).)).)))))).))))))).))))).......---- ( -33.00, z-score = -3.17, R) >droPer1.super_1 8496500 94 - 10282868 -------------UCUCUCUGUCUUAUUUCUUUUGGGGUAAGGAGAACAUGAUGUUCGACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGGAUCCAGAG---- -------------(((((.((((((((..(....)..)))).(((.((((((.((.((((((........)))))).)).)))))).))))))).))))).......---- ( -33.00, z-score = -3.17, R) >consensus ______UUUUUUAUGUUGUUGUUUUGUAU_UUUCGGGGCACCGAGAACAUGAUGUUCAACACCACAAUUUGUGUUGUACUUCGUGUACUCGACAUGGGCAAAUGGAGUGGG .............((((..((((...........((....))(((.((((((.((.((((((........)))))).)).)))))).)))))))..))))........... (-22.04 = -22.09 + 0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:30 2011