| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,828,040 – 14,828,168 |
| Length | 128 |
| Max. P | 0.973694 |

| Location | 14,828,040 – 14,828,168 |
|---|---|
| Length | 128 |
| Sequences | 7 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Shannon entropy | 0.51782 |
| G+C content | 0.50213 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
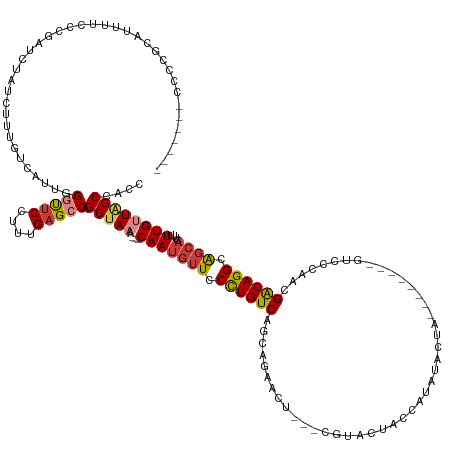
>dm3.chr3L 14828040 128 + 24543557 -------CCCCGCAUUUCCCCGAGCGAUCUUUGUCAUUGGGUUCCUUUGAGCCAGCUAAA---CAAUGUUCCCUGUCAGCAGAACU---CGUACUACCAUAUACUA--------GUCCCAACGACAGGCAGCACUUUUUGUUGGCCACC -------...........(((((..(((....))).)))))......((.((((((.(((---...((((.((((((....(.(((---.(((........))).)--------)).)....)))))).))))..))).)))))))).. ( -31.70, z-score = -1.69, R) >dp4.chrXR_group6 12399913 114 - 13314419 CGUCUUUCCCAAUUCUUUCCGCGUCUAUCGCUUUCGCUGGCUUCCUUUGAGCCAGCCAUAGUACAAUGUUCCCUGUCAGCAGUAGU---UGU----------------------------AGGACAGGCG----UUUUUGUUAGCCGCU ....................(((.(((..(((...((((((((.....))))))))...)))((((....(((((((.((((...)---)))----------------------------..)))))).)----...))))))).))). ( -32.40, z-score = -2.62, R) >droAna3.scaffold_13337 19010139 139 - 23293914 -------GGCCACGUCGUCUUCAUCUAUCCCUGUCAUUUGGUGCCUUUGAGCCAGCUAAA---CAAUGUUCCUUGUCAGCAGGACUACUCGUUCUAGUAGUUGUUCUUGCACUCCCUCCAAUGGCAGGCAGCAGUUUUUGCUAGCCACC -------(((...................(((((((((.(((((....((((..(((..(---(((......)))).)))..(((((((......)))))))))))..)))))......))))))))).(((((...))))).)))... ( -36.80, z-score = -1.06, R) >droYak2.chr3L 14911038 136 + 24197627 -------GCCCGCCUUUCCCCGAUCUAUCUUUGUCAUUGGGCUCCUUUGAGCCAGCUAAA---CAAUGUUCCCUGUCAGCAGAACU---CGUACUACCAUAUAGUAGUAUAGUGGUUCCACCGACAGGCAGCACUUUUUGUUAGCCGCC -------....((..........................(((((....))))).(((((.---(((((((.((((((.(..(((((---.(((((((......)))))))...)))))..).)))))).))))....)))))))).)). ( -39.00, z-score = -2.96, R) >droEre2.scaffold_4784 14787884 133 + 25762168 --------CCCGCCUCUCCCCGAUCUAUCUUUGCCAUUGG-UUCCUUUGAGCCAGCUAAA---CAAUGUUCCCUGUCAGCAGAACC---CGCACUACCAUAUACCA-UAUAGUAGUCCCAACGACAGGCAGCACUUUUUGUUAGCCGCC --------...((........................(((-(((....))))))(((((.---(((((((.((((((.((......---.))(((((.((((...)-))).)))))......)))))).))))....)))))))).)). ( -31.50, z-score = -3.12, R) >droSec1.super_0 6941702 131 + 21120651 ----UUCCCCCGCAUUUUCCCGAUCUAUCUUUGUCAUUGGGCUCCUUUGAGCCAGCUAAA---CAAUGUUCCCUGUCAGCAGAACU---CGUACUACCAUAUACUA--------GUCCCAACGACAGGCGGCACUUUUUGUUGGCCACC ----..............((((((...........))))))......((.((((((.(((---...(((..((((((....(.(((---.(((........))).)--------)).)....))))))..)))..))).)))))))).. ( -29.50, z-score = -1.10, R) >droSim1.chr3L 14143795 127 + 22553184 ----UUCCCCCGCAUUUUCCCGAUCUAUCUUUGUCAUUGGGCUCCUUUGAGCCAGCUAAA---CAAUGUUCCCUGUCAGCAGAACU---AGU----CAGCAUACUA--------GUCCCAACGACAGGCAGCACUUUUUGUUGGCCACC ----..............((((((...........))))))......((.((((((.(((---...((((.((((((....(.(((---(((----......))))--------)).)....)))))).))))..))).)))))))).. ( -32.40, z-score = -2.04, R) >consensus _______CCCCGCAUUUUCCCGAUCUAUCUUUGUCAUUGGGUUCCUUUGAGCCAGCUAAA___CAAUGUUCCCUGUCAGCAGAACU___CGUACUACCAUAUACUA________GUCCCAACGACAGGCAGCACUUUUUGUUAGCCACC .......................................(((((....))))).(((((.......((((.((((((.............................................)))))).)))).......))))).... (-17.04 = -17.45 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:57 2011