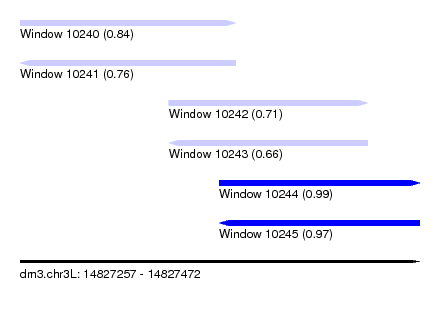
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,827,257 – 14,827,472 |
| Length | 215 |
| Max. P | 0.994649 |

| Location | 14,827,257 – 14,827,373 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.31200 |
| G+C content | 0.28759 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -11.70 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.836058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14827257 116 + 24543557 AAAUAAAUUUACAUUUGACCAUAAAAUGAUUUCAAACAGCAUUUAAGGCUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGAUAAAUGCUUGAUUAAAUCAUGUUUAUAUC--- ....................((((((((((((.....(((((((((((..(((((......))))))))..(((.....))).....)))))))).....))))))).)))))...--- ( -18.80, z-score = -2.05, R) >droYak2.chr3L 14910271 103 + 24197627 ----------AAAUGAGUUUAU---AUGAUUUCAAACAGCAUUUAACGUUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGAUAAAUGCUUGAUUAAAUCAUGUUUAUAUC--- ----------......((..((---(((((((.....((((((((.((..(((((......))))).....(((.....)))..)).)))))))).....)))))))))..))...--- ( -19.90, z-score = -2.73, R) >droEre2.scaffold_4784 14787094 119 + 25762168 AAAUAAAUUUUCAUUUACACAUUAUAUGAUUUCAAACAGCAUUUACCGCUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGGCAAAUGCUUGAUUAUCAUGUUUUUUUAUCAUA .......................(((((((.(((...(((((((.(((..(((((......))))).....(((.....)))..))).))))))))))..)))))))............ ( -20.40, z-score = -3.16, R) >droSec1.super_0 6940932 116 + 21120651 AAAUAAAUUUACAUUUGCCCAUAAAAUGAUGUCAAACAGCAUUUAAGGCUGAAUUUACAAUAAUUCCCGUCUGCUAUACGCACUCGAUAAAUGCUAGUUUAAAUCAUGUUCAUAUG--- ...................((((..(((((...((((((((((((((((.(((((......)))))..))))((.....))......)))))))).))))..))))).....))))--- ( -21.00, z-score = -1.91, R) >droSim1.chr3L 14143007 116 + 22553184 AAAUAAAUUUAUAUUUGCCCAUAAAAUGAUUUCAAACAGCAUUUAUGUCUGAAUUUACAAUAAUACCCGUCUGCUAUACGCACUCGAUAAAUGCUAGAUUAAAUCAUGUCUAUAUC--- .........................(((((((.....(((((((((((........)).........((..(((.....)))..))))))))))).....))))))).........--- ( -16.60, z-score = -1.35, R) >consensus AAAUAAAUUUACAUUUGCCCAUAAAAUGAUUUCAAACAGCAUUUAAGGCUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGAUAAAUGCUUGAUUAAAUCAUGUUUAUAUC___ .........................(((((((.....((((((((.....(((((......))))).....(((.....))).....)))))))).....)))))))............ (-11.70 = -12.58 + 0.88)

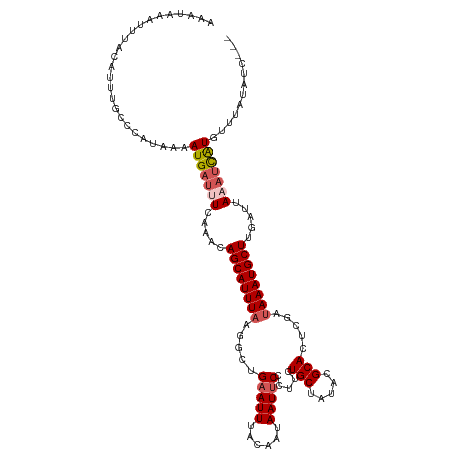
| Location | 14,827,257 – 14,827,373 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Shannon entropy | 0.31200 |
| G+C content | 0.28759 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -15.10 |
| Energy contribution | -17.02 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14827257 116 - 24543557 ---GAUAUAAACAUGAUUUAAUCAAGCAUUUAUCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAGCCUUAAAUGCUGUUUGAAAUCAUUUUAUGGUCAAAUGUAAAUUUAUUU ---((((((((.((((((...((((((((((...))))))..((((((.(((((((((......)))))..))))...))))))))))))))))))))).)))................ ( -25.00, z-score = -1.95, R) >droYak2.chr3L 14910271 103 - 24197627 ---GAUAUAAACAUGAUUUAAUCAAGCAUUUAUCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAACGUUAAAUGCUGUUUGAAAUCAU---AUAAACUCAUUU---------- ---.........(((((((.....((((((((.((..(((.....))).....(((((......)))))..)).)))))))).....)))))))---............---------- ( -22.20, z-score = -2.35, R) >droEre2.scaffold_4784 14787094 119 - 25762168 UAUGAUAAAAAAACAUGAUAAUCAAGCAUUUGCCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAGCGGUAAAUGCUGUUUGAAAUCAUAUAAUGUGUAAAUGAAAAUUUAUUU ((((((....((((.((.....))(((((((((((..(((.....))).....(((((......)))))..)))))))))))))))...))))))......((((((....)))))).. ( -29.70, z-score = -3.48, R) >droSec1.super_0 6940932 116 - 21120651 ---CAUAUGAACAUGAUUUAAACUAGCAUUUAUCGAGUGCGUAUAGCAGACGGGAAUUAUUGUAAAUUCAGCCUUAAAUGCUGUUUGACAUCAUUUUAUGGGCAAAUGUAAAUUUAUUU ---..((((((.(((((((((((.(((((((((((..(((.....)))..)))(((((......))))).....)))))))))))))).)))))))))))................... ( -26.20, z-score = -1.75, R) >droSim1.chr3L 14143007 116 - 22553184 ---GAUAUAGACAUGAUUUAAUCUAGCAUUUAUCGAGUGCGUAUAGCAGACGGGUAUUAUUGUAAAUUCAGACAUAAAUGCUGUUUGAAAUCAUUUUAUGGGCAAAUAUAAAUUUAUUU ---..((((((.(((((((....((((((((((((..(((.....)))..))).......(((........))))))))))))....)))))))))))))................... ( -22.30, z-score = -1.02, R) >consensus ___GAUAUAAACAUGAUUUAAUCAAGCAUUUAUCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAGCCUUAAAUGCUGUUUGAAAUCAUUUUAUGGGCAAAUGUAAAUUUAUUU .....((((((.(((((((.....((((((((((...(((.....)))...))(((((......))))).....)))))))).....)))))))))))))................... (-15.10 = -17.02 + 1.92)

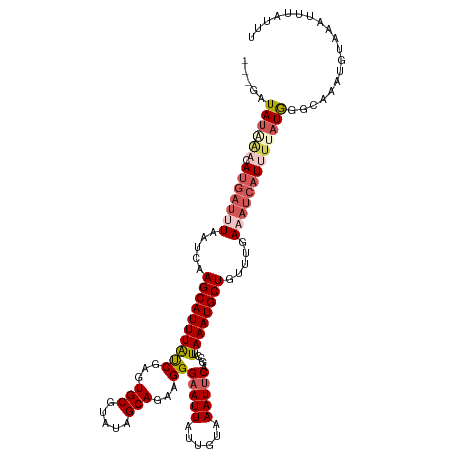
| Location | 14,827,337 – 14,827,444 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.08 |
| Shannon entropy | 0.41934 |
| G+C content | 0.32302 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -9.33 |
| Energy contribution | -11.00 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
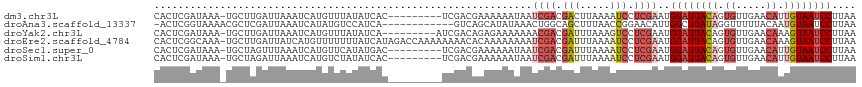
>dm3.chr3L 14827337 107 + 24543557 CACUCGAUAAA-UGCUUGAUUAAAUCAUGUUUAUAUCAC---------UCGACGAAAAAAUAAUCGACGACUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAA ...((((((..-.((.((((...)))).))...))))..---------(((.(((........))).)))............))..(((((((((((.....))))))))))).... ( -23.60, z-score = -3.15, R) >droAna3.scaffold_13337 19009353 104 - 23293914 -ACUCGGUAAAACGCUCGAUUAAAUCAUAUGUCCAUCA------------GUCAGCAUAUAAACUGGCAGCUUUAACCGGAACAUUGGCUUAUAGGUUUUUACAAUGUAAUCCUUAA -....(((.(((.(((.(((...)))............------------(((((........))))))))))).)))(((((((((...............))))))..))).... ( -18.86, z-score = -0.71, R) >droYak2.chr3L 14910338 107 + 24197627 CACUCGAUAAA-UGCUUGAUUAAAUCAUGUUUAUAUCA---------AUCGACAGAGAAAAAAACGACGAUUUAAAGUCCUCGAAUGGAUUACAGUGUUGAACAAAGUAAUCCUUAA .....((((..-.((.((((...)))).))...)))).---------.................(((.(((.....))).)))...(((((((..((.....))..))))))).... ( -19.00, z-score = -1.02, R) >droEre2.scaffold_4784 14787174 116 + 25762168 CACUCGGCAAA-UGCUUGAUUAUCAUGUUUUUUUAUCAUAGACCAAAAAAAACACAAAAAAAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAAAGUAAUCCUUAA ...(((((...-.)).)))......(((((((((...........))))))))).........((((.(((.....))).))))..(((((((..((.....))..))))))).... ( -20.70, z-score = -1.89, R) >droSec1.super_0 6941012 107 + 21120651 CACUCGAUAAA-UGCUAGUUUAAAUCAUGUUCAUAUGAC---------UCGACGAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAA ...((((((((-(....)))))..(((((....))))).---------))))...........((((.(((.....))).))))..(((((((((((.....))))))))))).... ( -24.60, z-score = -2.98, R) >droSim1.chr3L 14143087 107 + 22553184 CACUCGAUAAA-UGCUAGAUUAAAUCAUGUCUAUAUCAC---------UCGACGAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAA ...((((...(-((.(((((........))))))))...---------))))...........((((.(((.....))).))))..(((((((((((.....))))))))))).... ( -25.80, z-score = -3.77, R) >consensus CACUCGAUAAA_UGCUUGAUUAAAUCAUGUUUAUAUCAC_________UCGACAAAAAAAAAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAAUGUAAUCCUUAA ...............................................................((((.(((.....))).))))..((((((((.((.....)).)))))))).... ( -9.33 = -11.00 + 1.67)
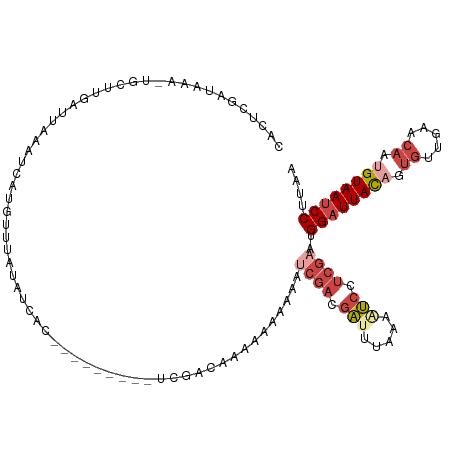
| Location | 14,827,337 – 14,827,444 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Shannon entropy | 0.41934 |
| G+C content | 0.32302 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -11.30 |
| Energy contribution | -13.00 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14827337 107 - 24543557 UUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAGUCGUCGAUUAUUUUUUCGUCGA---------GUGAUAUAAACAUGAUUUAAUCAAGCA-UUUAUCGAGUG ....((((((((.((.....)).))))))))((((((..........(((.(((........))).)))---------.((((.((((.....))))))))....-....)))))). ( -24.70, z-score = -2.26, R) >droAna3.scaffold_13337 19009353 104 + 23293914 UUAAGGAUUACAUUGUAAAAACCUAUAAGCCAAUGUUCCGGUUAAAGCUGCCAGUUUAUAUGCUGAC------------UGAUGGACAUAUGAUUUAAUCGAGCGUUUUACCGAGU- ....(((..((((((...............)))))))))(((.(((((.((..(.(((((((((...------------....)).)))))))......)..))))))))))....- ( -18.36, z-score = 0.13, R) >droYak2.chr3L 14910338 107 - 24197627 UUAAGGAUUACUUUGUUCAACACUGUAAUCCAUUCGAGGACUUUAAAUCGUCGUUUUUUUCUCUGUCGAU---------UGAUAUAAACAUGAUUUAAUCAAGCA-UUUAUCGAGUG ....(((((((..((.....))..)))))))((((((.(...(((((((((.((((...((.........---------.))...))))))))))))).....).-....)))))). ( -24.10, z-score = -2.36, R) >droEre2.scaffold_4784 14787174 116 - 25762168 UUAAGGAUUACUUUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUUUUUUUGUGUUUUUUUUGGUCUAUGAUAAAAAAACAUGAUAAUCAAGCA-UUUGCCGAGUG ....(((((((..((.....))..)))))))((((((.(((.....))).))((((....(..(((((((((..(((...))))))))))))..).))))..((.-...)).)))). ( -26.30, z-score = -2.22, R) >droSec1.super_0 6941012 107 - 21120651 UUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGA---------GUCAUAUGAACAUGAUUUAAACUAGCA-UUUAUCGAGUG ....((((((((.((.....)).))))))))((((((..........(((.(((........))).)))---------(((((......)))))...........-....)))))). ( -25.80, z-score = -2.68, R) >droSim1.chr3L 14143087 107 - 22553184 UUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGA---------GUGAUAUAGACAUGAUUUAAUCUAGCA-UUUAUCGAGUG ....((((((((.((.....)).))))))))..((((.(((.....))).)))).......((((..((---------(((...((((..........)))).))-)))..)))).. ( -26.80, z-score = -2.70, R) >consensus UUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGA_________GUGAUAUAAACAUGAUUUAAUCAAGCA_UUUAUCGAGUG ....((((((((.((.....)).))))))))..((((.(((.....))).))))............................................................... (-11.30 = -13.00 + 1.70)
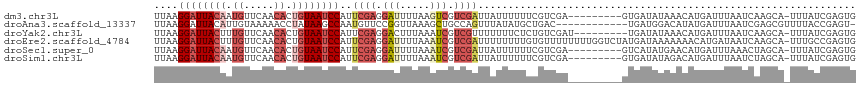
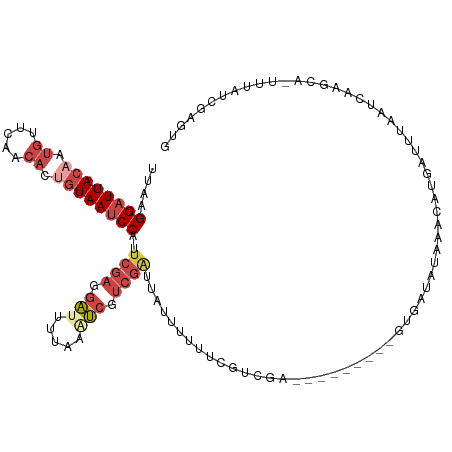
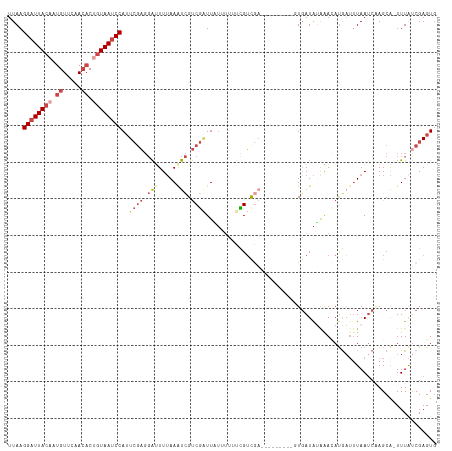
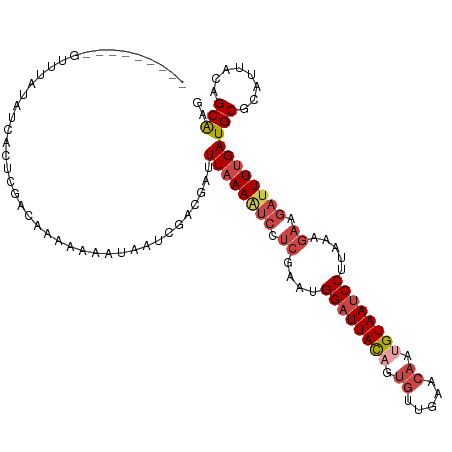
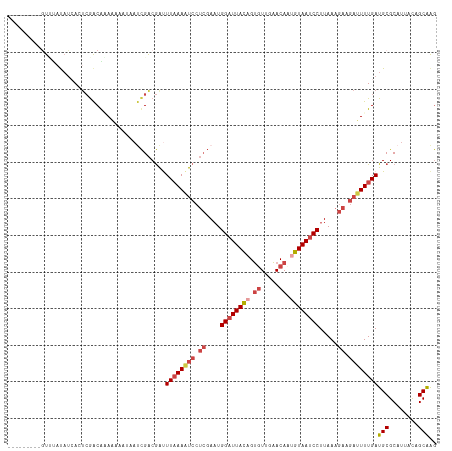
| Location | 14,827,364 – 14,827,472 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Shannon entropy | 0.38208 |
| G+C content | 0.34016 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -15.95 |
| Energy contribution | -17.40 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.994649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14827364 108 + 24543557 ---------GUUUAUAUCACUCGACGAAAAAAUAAUCGACGACUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAG ---------...........(((.(((........))).))).((((((((.((....(((((((((((.....))))))))))).....)).))))))))(((........))).. ( -28.90, z-score = -4.30, R) >droAna3.scaffold_13337 19009380 105 - 23293914 ---------AUGUCCAUCAGUCAGCAUAUAAA---CUGGCAGCUUUAACCGGAACAUUGGCUUAUAGGUUUUUACAAUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUAUAGCGGA ---------((((.((((((((((........---)))))..((((((..(((((((((...............))))))..))))))))).......))))).))))......... ( -23.26, z-score = -0.80, R) >droYak2.chr3L 14910365 108 + 24197627 ---------GUUUAUAUCAAUCGACAGAGAAAAAAACGACGAUUUAAAGUCCUCGAAUGGAUUACAGUGUUGAACAAAGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAG ---------((((.......((....)).....))))......((((((((.((....(((((((..((.....))..))))))).....)).))))))))(((........))).. ( -21.20, z-score = -1.66, R) >droEre2.scaffold_4784 14787201 117 + 25762168 UUUUUUAUCAUAGACCAAAAAAAACACAAAAAAAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAAAGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAG ...........................................((((((((.((....(((((((..((.....))..))))))).....)).))))))))(((........))).. ( -20.50, z-score = -2.36, R) >droSec1.super_0 6941039 108 + 21120651 ---------GUUCAUAUGACUCGACGAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAG ---------...........(((.(((........))).))).((((((((.((....(((((((((((.....))))))))))).....)).))))))))(((........))).. ( -29.10, z-score = -3.97, R) >droSim1.chr3L 14143114 108 + 22553184 ---------GUCUAUAUCACUCGACGAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAG ---------...........(((.(((........))).))).((((((((.((....(((((((((((.....))))))))))).....)).))))))))(((........))).. ( -29.10, z-score = -4.21, R) >consensus _________GUUUAUAUCACUCGACAAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAAUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAG ...........................................((((((((.((....((((((((.((.....)).)))))))).....)).))))))))(((........))).. (-15.95 = -17.40 + 1.45)
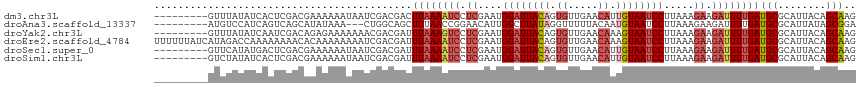
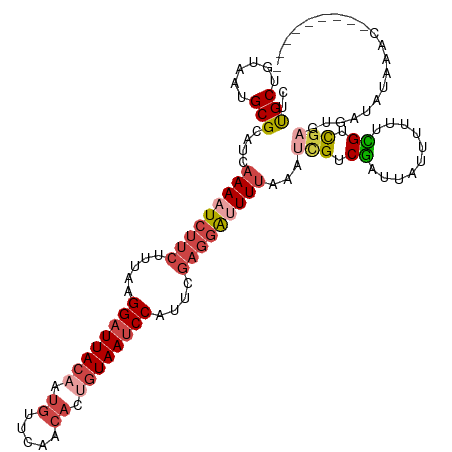
| Location | 14,827,364 – 14,827,472 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Shannon entropy | 0.38208 |
| G+C content | 0.34016 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -14.31 |
| Energy contribution | -15.98 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14827364 108 - 24543557 CUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAGUCGUCGAUUAUUUUUUCGUCGAGUGAUAUAAAC--------- ...((......))..((((((((((((.....((((((((.((.....)).))))))))....))))))))).(.(((.(((........))).))).))))......--------- ( -28.40, z-score = -3.52, R) >droAna3.scaffold_13337 19009380 105 + 23293914 UCCGCUAUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAUUGUAAAAACCUAUAAGCCAAUGUUCCGGUUAAAGCUGCCAG---UUUAUAUGCUGACUGAUGGACAU--------- ..(((......)))(((((.......(((((.(((..((((((...............)))))))))...)))))....(((---(......))))..))))).....--------- ( -19.36, z-score = -0.31, R) >droYak2.chr3L 14910365 108 - 24197627 CUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACUUUGUUCAACACUGUAAUCCAUUCGAGGACUUUAAAUCGUCGUUUUUUUCUCUGUCGAUUGAUAUAAAC--------- ...((......))..((((((.(((((.....(((((((..((.....))..)))))))....))))).))).(((((.((..........)).))))))))......--------- ( -19.90, z-score = -1.52, R) >droEre2.scaffold_4784 14787201 117 - 25762168 CUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACUUUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUUUUUUUGUGUUUUUUUUGGUCUAUGAUAAAAAA ...((((.((.(((((..((((((........(((((((..((.....))..)))))))....((.(((.....))).))))))))...)))))...)).))))............. ( -23.40, z-score = -2.06, R) >droSec1.super_0 6941039 108 - 21120651 CUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGAGUCAUAUGAAC--------- ...((......))...(((((((((((.....((((((((.((.....)).))))))))....)))))))))...(((.(((........))).))).......))..--------- ( -27.10, z-score = -3.25, R) >droSim1.chr3L 14143114 108 - 22553184 CUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGAGUGAUAUAGAC--------- ....(((((...(((...(((((((((.....((((((((.((.....)).))))))))....)))))))))...(((.(((........))).)))))).)))))..--------- ( -31.30, z-score = -4.31, R) >consensus CUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGAGUGAUAUAAAC_________ ..(((......)))....(((((((((.....((((((((.((.....)).))))))))....)))))))))...(((.((..........)).))).................... (-14.31 = -15.98 + 1.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:56 2011