
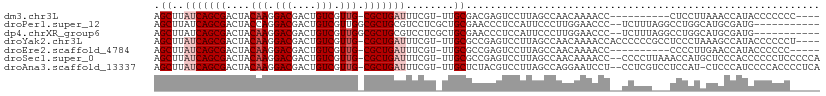
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,826,407 – 14,826,502 |
| Length | 95 |
| Max. P | 0.795623 |

| Location | 14,826,407 – 14,826,502 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.96 |
| Shannon entropy | 0.56537 |
| G+C content | 0.55738 |
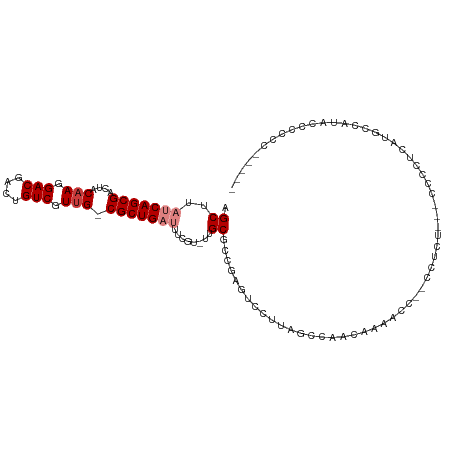
| Mean single sequence MFE | -21.79 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14826407 95 + 24543557 AGCUUAUCAGCGACUACAAGGACGACUGUCGUUG-CGCUGAUUUCGU-UUGCGACGAGUCCUUAGCCAACAAAACC----------CUCCUUAAACCAUACCCCCCC---- .(((....)))......(((((.(.....)((((-.(((((.(((((-(...))))))...)))))))))......----------.)))))...............---- ( -18.90, z-score = -1.44, R) >droPer1.super_12 818743 98 - 2414086 AGCUUAUCAGCGACUACCAGGACGACUGUCGUUGGCGCUGCGUCCUCGCUGCGAACCCUCCAUUCCCUUGGAACCC--UCUUUAGGCCUGGCAUGCGAUG----------- ....((((.(((....(((((....(((..(..(((((.(((....))).))).....((((......)))).)).--.)..))).)))))..)))))))----------- ( -27.40, z-score = -0.05, R) >dp4.chrXR_group6 12398496 98 - 13314419 AGCUUAUCAGCGACUACAAGGACGACUGUCGUUGGCGCUGCGUCCUCGCUGCGAACCCUCCAUUCCCUUGGAACCC--UCUUUAGGCCUGGCAUGCGAUG----------- ....((((((((....(((.(((....))).))).))))((((((..(((..((....((((......))))....--))....)))..)).))))))))----------- ( -27.30, z-score = -0.06, R) >droYak2.chr3L 14909341 105 + 24197627 AGCUUAUCAGCGACUACAAGGACGACUGUCGUUG-CGCUGAUUUCGU-UUGCGCCGAGUCCUUAGCCAACAAAACCACCCCCCGCCUCCCUAAAGCCAUACCCCCCU---- .(((((((((((....(((.(((....))).)))-)))))))...((-(.((............)).)))......................))))...........---- ( -20.00, z-score = -1.13, R) >droEre2.scaffold_4784 14786130 94 + 25762168 AGCUUAUCAGCGACUACAAGGACGACUGUCGUUG-CGCUGAUUUCGU-UUGCGCCGAGUCCUUAGCCAACAAAACC----------CCCCUUGAACCAUACCCCCC----- .(((....))).....(((((((....)))((((-.(((((.((((.-......))))...)))))))))......----------...)))).............----- ( -19.00, z-score = -1.35, R) >droSec1.super_0 6940081 107 + 21120651 AGCUUAUCAGCGACUACAAGGACGACUGUCGUUG-CGCUGAUUUCGU-UUGCGCCGAGUCCUUAGCCAACAAAACC--CCCCUUAAACCAUGCUCCCACCCCCCUCCCCCA (((..(((((((....(((.(((....))).)))-)))))))...((-(.((............)).)))......--.............)))................. ( -18.40, z-score = -1.13, R) >droAna3.scaffold_13337 19008462 106 - 23293914 AGCUUAUCAGCGACUACAAGGACGACUGUCGUUG-CGCUGAUUUCGU-UUGCUCUACGUCCUUAGCCAGGAAUCCU--CCUCGUCCUCCAU-CUCCCAUCCCCACCCCUCA .(((....))).......(((((((.......((-.(((((...(((-.......)))...)))))))(((....)--)))))))))....-................... ( -21.50, z-score = -1.10, R) >consensus AGCUUAUCAGCGACUACAAGGACGACUGUCGUUG_CGCUGAUUUCGU_UUGCGCCGAGUCCUUAGCCAACAAAACC__CCUCU___CCCCUCAUGCCAUACCCCCC_____ .((..(((((((....(((.(((....))).))).)))))))........))........................................................... (-13.16 = -13.87 + 0.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:50 2011