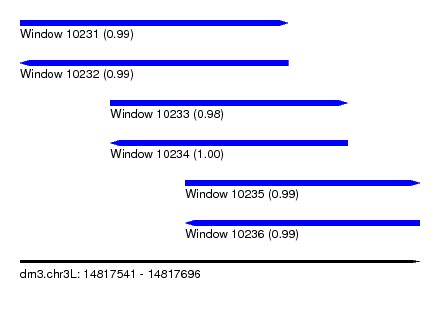
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,817,541 – 14,817,696 |
| Length | 155 |
| Max. P | 0.998595 |

| Location | 14,817,541 – 14,817,645 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Shannon entropy | 0.49029 |
| G+C content | 0.49280 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -20.57 |
| Energy contribution | -22.86 |
| Covariance contribution | 2.29 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.992701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
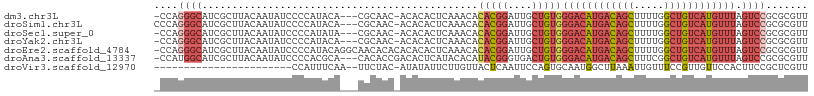
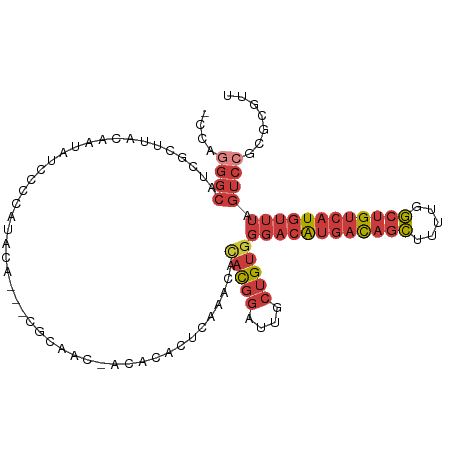
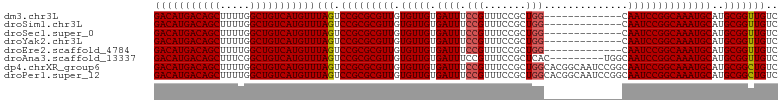
>dm3.chr3L 14817541 104 + 24543557 -CCAGGGCAUCGCUUACAAUAUCCCCAUACA---CGCAAC-ACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUU -...((((...............((((((..---.((((.-.(..............)..))))))))))((((((((((.....))))))))))...))))....... ( -32.34, z-score = -2.43, R) >droSim1.chr3L 14140307 105 + 22553184 CCCAGGGCAUCGCUUACAAUAUCCCCAUACA---CGCAAC-ACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUU ....((((...............((((((..---.((((.-.(..............)..))))))))))((((((((((.....))))))))))...))))....... ( -32.34, z-score = -2.24, R) >droSec1.super_0 6938925 104 + 21120651 -CCAGGGCAUCGCUUACAAUAUCCCCAUAUA---CGCAAC-ACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUU -...((((...............((((((..---.((((.-.(..............)..))))))))))((((((((((.....))))))))))...))))....... ( -32.34, z-score = -2.46, R) >droYak2.chr3L 14908141 104 + 24197627 -CCAGGGCAUCGCUUACAAUAUCCCCAUACA---CGCAAC-ACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUU -...((((...............((((((..---.((((.-.(..............)..))))))))))((((((((((.....))))))))))...))))....... ( -32.34, z-score = -2.43, R) >droEre2.scaffold_4784 14784896 108 + 25762168 -CCAGGGCAUCGCUUACAAUAUCCCCAUACAGGCAACACACACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUU -...((((...............(((((...(((((..(..................)..))))))))))((((((((((.....))))))))))...))))....... ( -32.67, z-score = -2.12, R) >droAna3.scaffold_13337 19007460 105 - 23293914 -CCAUGGCAUCGCUUACAAUAUCCCCACGCA---CACACCGACACUCAUACACAUACGGGUGACUGUGGGACAUGACAGCUUUCGGCUGUCAUGUUUAGUCCGCGCGUU -.........(((..((......((((((..---(((.(((...............))))))..))))))((((((((((.....))))))))))...))..))).... ( -34.86, z-score = -2.52, R) >droVir3.scaffold_12970 10022230 83 - 11907090 -----------------------CCAUUUCAA--UUCUAC-AUAUAUUCUUGUUACUCAAUUCCAGUGCAAUGGCUUAAAUUGUUUCCGUUGUUCCACUUCCGCUCGUU -----------------------.........--......-.......................((((((((((............))))))...)))).......... ( -6.00, z-score = 0.11, R) >consensus _CCAGGGCAUCGCUUACAAUAUCCCCAUACA___CGCAAC_ACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUU ....((((..............................................(((((....)))))((((((((((((.....)))))))))))).))))....... (-20.57 = -22.86 + 2.29)
| Location | 14,817,541 – 14,817,645 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Shannon entropy | 0.49029 |
| G+C content | 0.49280 |
| Mean single sequence MFE | -33.99 |
| Consensus MFE | -24.05 |
| Energy contribution | -25.80 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14817541 104 - 24543557 AACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGU-GUUGCG---UGUAUGGGGAUAUUGUAAGCGAUGCCCUGG- .((((((((.....((((((((((.....))))))))))..((((.(((.........)))..)))).-.)))))---)))..((((.(((((....)))))))))..- ( -37.80, z-score = -2.23, R) >droSim1.chr3L 14140307 105 - 22553184 AACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGU-GUUGCG---UGUAUGGGGAUAUUGUAAGCGAUGCCCUGGG .((((((((.....((((((((((.....))))))))))..((((.(((.........)))..)))).-.)))))---)))..((((.(((((....)))))))))... ( -37.80, z-score = -1.87, R) >droSec1.super_0 6938925 104 - 21120651 AACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGU-GUUGCG---UAUAUGGGGAUAUUGUAAGCGAUGCCCUGG- ((((((((((((..((((((((((.....)))))))))).....))...)))))))))).........-......---.....((((.(((((....)))))))))..- ( -36.20, z-score = -1.99, R) >droYak2.chr3L 14908141 104 - 24197627 AACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGU-GUUGCG---UGUAUGGGGAUAUUGUAAGCGAUGCCCUGG- .((((((((.....((((((((((.....))))))))))..((((.(((.........)))..)))).-.)))))---)))..((((.(((((....)))))))))..- ( -37.80, z-score = -2.23, R) >droEre2.scaffold_4784 14784896 108 - 25762168 AACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGUGUGUGUUGCCUGUAUGGGGAUAUUGUAAGCGAUGCCCUGG- .(((((((.(((..((((((((((.....))))))))))..((((........))))....))).)))))))...........((((.(((((....)))))))))..- ( -38.70, z-score = -2.21, R) >droAna3.scaffold_13337 19007460 105 + 23293914 AACGCGCGGACUAAACAUGACAGCCGAAAGCUGUCAUGUCCCACAGUCACCCGUAUGUGUAUGAGUGUCGGUGUG---UGCGUGGGGAUAUUGUAAGCGAUGCCAUGG- .(((((((.(((..(((((((((.(....))))))))))......(.(((.((((....)))).))).)))).))---)))))..((.(((((....)))))))....- ( -40.60, z-score = -2.25, R) >droVir3.scaffold_12970 10022230 83 + 11907090 AACGAGCGGAAGUGGAACAACGGAAACAAUUUAAGCCAUUGCACUGGAAUUGAGUAACAAGAAUAUAU-GUAGAA--UUGAAAUGG----------------------- ..(((((....))...(((..(....).((((((.(((......)))..))))))............)-))....--)))......----------------------- ( -9.00, z-score = 0.42, R) >consensus AACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGU_GUUGCG___UGUAUGGGGAUAUUGUAAGCGAUGCCCUGG_ .(((((((((((..((((((((((.....)))))))))).....))...))))))))).........................((((.(((((....)))))))))... (-24.05 = -25.80 + 1.75)

| Location | 14,817,576 – 14,817,668 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.66 |
| Shannon entropy | 0.10052 |
| G+C content | 0.50258 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -30.20 |
| Energy contribution | -30.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14817576 92 + 24543557 -CACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCU---- -(((((((...(((.(.(((((((......(((((((((((.....)))))))))))))))))).).)))))).))))...............---- ( -33.30, z-score = -2.57, R) >droSim1.chr3L 14140343 92 + 22553184 -CACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCU---- -(((((((...(((.(.(((((((......(((((((((((.....)))))))))))))))))).).)))))).))))...............---- ( -33.30, z-score = -2.57, R) >droSec1.super_0 6938960 92 + 21120651 -CACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCU---- -(((((((...(((.(.(((((((......(((((((((((.....)))))))))))))))))).).)))))).))))...............---- ( -33.30, z-score = -2.57, R) >droYak2.chr3L 14908176 92 + 24197627 -CACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCU---- -(((((((...(((.(.(((((((......(((((((((((.....)))))))))))))))))).).)))))).))))...............---- ( -33.30, z-score = -2.57, R) >droEre2.scaffold_4784 14784935 92 + 25762168 -CACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCU---- -(((((((...(((.(.(((((((......(((((((((((.....)))))))))))))))))).).)))))).))))...............---- ( -33.30, z-score = -2.57, R) >droAna3.scaffold_13337 19007495 97 - 23293914 CGACACUCAUACACAUACGGGUGACUGUGGGACAUGACAGCUUUCGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUCACU ...(((....(((((.(((.(((((((...(((((((((((.....))))))))))))))).))).)))))))).)))................... ( -36.10, z-score = -2.67, R) >consensus _CACACACUCAAACACACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCU____ .(((((((...(((.(.(((((((......(((((((((((.....)))))))))))))))))).).)))))).))))................... (-30.20 = -30.45 + 0.25)


| Location | 14,817,576 – 14,817,668 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 94.66 |
| Shannon entropy | 0.10052 |
| G+C content | 0.50258 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -33.40 |
| Energy contribution | -34.23 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.11 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14817576 92 - 24543557 ----AGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGUG- ----..((....)).....((((.(((((((((((((((..((((((((((.....)))))))))).....))...))))))))))...)))))))- ( -36.60, z-score = -4.21, R) >droSim1.chr3L 14140343 92 - 22553184 ----AGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGUG- ----..((....)).....((((.(((((((((((((((..((((((((((.....)))))))))).....))...))))))))))...)))))))- ( -36.60, z-score = -4.21, R) >droSec1.super_0 6938960 92 - 21120651 ----AGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGUG- ----..((....)).....((((.(((((((((((((((..((((((((((.....)))))))))).....))...))))))))))...)))))))- ( -36.60, z-score = -4.21, R) >droYak2.chr3L 14908176 92 - 24197627 ----AGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGUG- ----..((....)).....((((.(((((((((((((((..((((((((((.....)))))))))).....))...))))))))))...)))))))- ( -36.60, z-score = -4.21, R) >droEre2.scaffold_4784 14784935 92 - 25762168 ----AGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGUG- ----..((....)).....((((.(((((((((((((((..((((((((((.....)))))))))).....))...))))))))))...)))))))- ( -36.60, z-score = -4.21, R) >droAna3.scaffold_13337 19007495 97 + 23293914 AGUGAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCGAAAGCUGUCAUGUCCCACAGUCACCCGUAUGUGUAUGAGUGUCG .((((.((....))....)))).((((((((.(..((((..(((((((((.(....)))))))))).....))))..)))).))))).......... ( -35.90, z-score = -3.59, R) >consensus ____AGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGUGUGUUUGAGUGUGUG_ ......((....)).....((((.(((((((((((((((..((((((((((.....)))))))))).....))...))))))))))...))))))). (-33.40 = -34.23 + 0.83)


| Location | 14,817,605 – 14,817,696 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Shannon entropy | 0.13371 |
| G+C content | 0.52561 |
| Mean single sequence MFE | -38.29 |
| Consensus MFE | -31.58 |
| Energy contribution | -31.39 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.991925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14817605 91 + 24543557 GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGG-------------CAAUCCGGCAAAUGCAUGCGGUUGUC (((((((((((.....)))))))))))....(((((((((.(((((.((((.(((.......)))-------------.))))))))))))))..))))..... ( -36.40, z-score = -3.41, R) >droSim1.chr3L 14140372 91 + 22553184 GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGG-------------CAAUCCGGCAAAUGCAUGCGGUUGUC (((((((((((.....)))))))))))....(((((((((.(((((.((((.(((.......)))-------------.))))))))))))))..))))..... ( -36.40, z-score = -3.41, R) >droSec1.super_0 6938989 91 + 21120651 GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGG-------------CAAUCCGGCAAAUGCAUGCGGUUGUC (((((((((((.....)))))))))))....(((((((((.(((((.((((.(((.......)))-------------.))))))))))))))..))))..... ( -36.40, z-score = -3.41, R) >droYak2.chr3L 14908205 91 + 24197627 GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGG-------------CAAUCCGGCAAAUGCAUGCGGUUGUC (((((((((((.....)))))))))))....(((((((((.(((((.((((.(((.......)))-------------.))))))))))))))..))))..... ( -36.40, z-score = -3.41, R) >droEre2.scaffold_4784 14784964 91 + 25762168 GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGG-------------CAAUCCGGCAAAUGCAUGCGGUUGUC (((((((((((.....)))))))))))....(((((((((.(((((.((((.(((.......)))-------------.))))))))))))))..))))..... ( -36.40, z-score = -3.41, R) >droAna3.scaffold_13337 19007525 95 - 23293914 GACAUGACAGCUUUCGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUCAC---------UGGCAAUCCGGCAAAUGCAUGCGGUUGUC (((((((((((.....)))))))))))....(((((((((.(((((.((((.((((........))---------.)).))))))))))))))..))))..... ( -35.90, z-score = -2.80, R) >dp4.chrXR_group6 12397097 104 - 13314419 GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGGCACGGCAAUCCGGCAAUCCGGCAAAUGCAUGCGGCUGUC (((((((((((.....)))))))))))(((.(((((((((.(((((.((((.(((..(..((((...)))).)..))).))))))))))))))..))))))).. ( -44.20, z-score = -3.13, R) >droPer1.super_12 817369 104 - 2414086 GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGGCACGGCAAUCCGGCAAUCCGGCAAAUGCAUGCGGCUGUC (((((((((((.....)))))))))))(((.(((((((((.(((((.((((.(((..(..((((...)))).)..))).))))))))))))))..))))))).. ( -44.20, z-score = -3.13, R) >consensus GACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUGG_____________CAAUCCGGCAAAUGCAUGCGGUUGUC (((((((((((.....)))))))))))(((.(((((((((.(((((.((((............................))))))))))))))..))))))).. (-31.58 = -31.39 + -0.19)
| Location | 14,817,605 – 14,817,696 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.58 |
| Shannon entropy | 0.13371 |
| G+C content | 0.52561 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
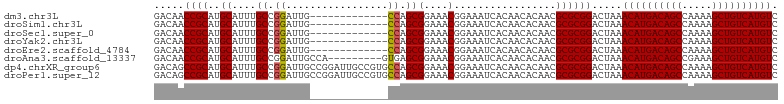
>dm3.chr3L 14817605 91 - 24543557 GACAACCGCAUGCAUUUGCCGGAUUG-------------CCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC .....((((..((....((.((....-------------)).))(....).................)))))).....((((((((((.....)))))))))). ( -29.20, z-score = -3.09, R) >droSim1.chr3L 14140372 91 - 22553184 GACAACCGCAUGCAUUUGCCGGAUUG-------------CCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC .....((((..((....((.((....-------------)).))(....).................)))))).....((((((((((.....)))))))))). ( -29.20, z-score = -3.09, R) >droSec1.super_0 6938989 91 - 21120651 GACAACCGCAUGCAUUUGCCGGAUUG-------------CCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC .....((((..((....((.((....-------------)).))(....).................)))))).....((((((((((.....)))))))))). ( -29.20, z-score = -3.09, R) >droYak2.chr3L 14908205 91 - 24197627 GACAACCGCAUGCAUUUGCCGGAUUG-------------CCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC .....((((..((....((.((....-------------)).))(....).................)))))).....((((((((((.....)))))))))). ( -29.20, z-score = -3.09, R) >droEre2.scaffold_4784 14784964 91 - 25762168 GACAACCGCAUGCAUUUGCCGGAUUG-------------CCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC .....((((..((....((.((....-------------)).))(....).................)))))).....((((((((((.....)))))))))). ( -29.20, z-score = -3.09, R) >droAna3.scaffold_13337 19007525 95 + 23293914 GACAACCGCAUGCAUUUGCCGGAUUGCCA---------GUGAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCGAAAGCUGUCAUGUC .....((((..((.......((....)).---------((((.((....))....))))........)))))).....(((((((((.(....)))))))))). ( -33.00, z-score = -3.30, R) >dp4.chrXR_group6 12397097 104 + 13314419 GACAGCCGCAUGCAUUUGCCGGAUUGCCGGAUUGCCGUGCCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC ...((((((..(((....((((....))))..))).(((....((....)).....)))..........)))).))..((((((((((.....)))))))))). ( -38.40, z-score = -2.90, R) >droPer1.super_12 817369 104 + 2414086 GACAGCCGCAUGCAUUUGCCGGAUUGCCGGAUUGCCGUGCCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC ...((((((..(((....((((....))))..))).(((....((....)).....)))..........)))).))..((((((((((.....)))))))))). ( -38.40, z-score = -2.90, R) >consensus GACAACCGCAUGCAUUUGCCGGAUUG_____________CCAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUC .....((((..((....))........................((....))..................)))).....((((((((((.....)))))))))). (-27.12 = -27.12 + 0.00)
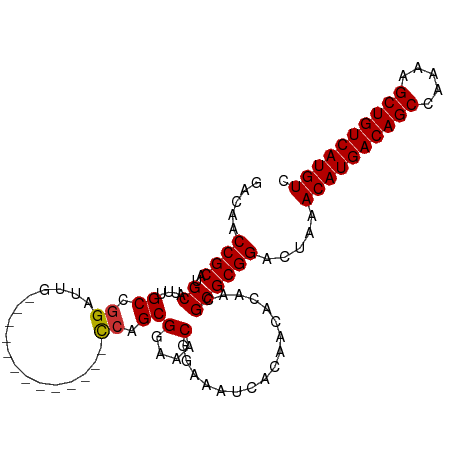
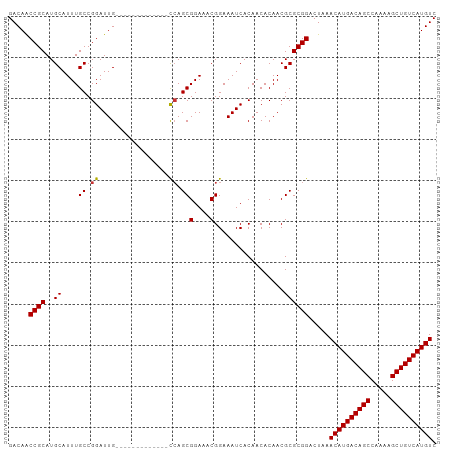
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:50 2011