| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,804,134 – 14,804,226 |
| Length | 92 |
| Max. P | 0.734932 |

| Location | 14,804,134 – 14,804,226 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Shannon entropy | 0.33882 |
| G+C content | 0.42737 |
| Mean single sequence MFE | -20.15 |
| Consensus MFE | -14.99 |
| Energy contribution | -14.99 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
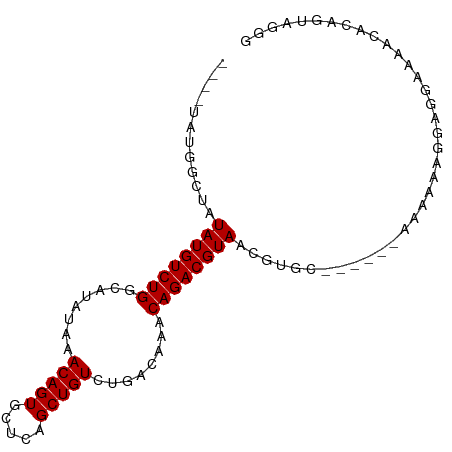
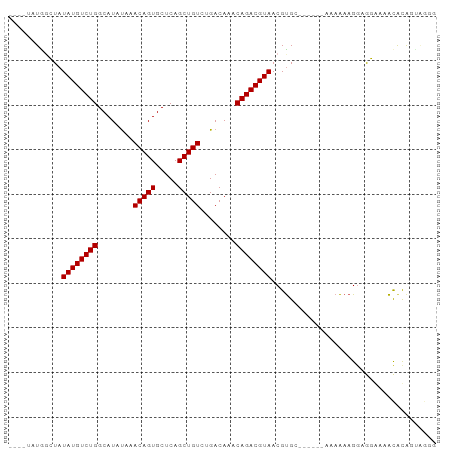
>dm3.chr3L 14804134 92 - 24543557 ----UAUGGCUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCUGACAAACAGACGUAACGUGCAAAAAAAAAAAAGGAGGAAAACAUAGCAGGG ----....((((((((((((((((.......))))((((....))))....)))))))).....................(.....).)))).... ( -18.00, z-score = -0.99, R) >droSim1.chr3L_random 719646 90 - 1049610 ----UAUGGCUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCUGACAAACAGACGUAACGUGC--AAAAAAAAAAGGAGGAAAACGUAGUAGGG ----....(((.((((((((((((.......))))((((....))))....))))))))((((..--..................))))))).... ( -17.45, z-score = -0.61, R) >droSec1.super_0 6925237 91 - 21120651 ----UAUGGCUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCUGACAAACAGACGUAACGUGC-AAAAAAAAAAAGGAGGAAAACGUAGUAGGG ----....(((.((((((((((((.......))))((((....))))....))))))))((((..-...................))))))).... ( -17.40, z-score = -0.63, R) >droYak2.chr3L 14892782 90 - 24197627 UAUGGCUGGCUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCUGACAAACAGACGUAACGUGC------AAAAAAGGAGGGGUACACAGUGGGG ..(.((((..((((((.....))))))....((((((..(((((((.....)))))((.....))------......))..)))))).)))).).. ( -24.10, z-score = -1.07, R) >droEre2.scaffold_4784 14771312 86 - 25762168 ----UAUGGCUAUAUGUCUGGCAUACAAACAGUGCUCAGCUGUCUGACAAACAGACGUAACGCGG------AAAGCAGGAGGGGAGCACAGUAGGG ----.....((((.(((.((.....)).)))((((((..(((((((.....))))).....((..------...))...))..)))))).)))).. ( -24.50, z-score = -1.59, R) >droAna3.scaffold_13337 18994960 83 + 23293914 ----GGUAGGUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCAGACAAACAGACGUAACGUG-------CAAAAAGAUACGAAAUACAGUAG-- ----.(((.((.((((((((........(((((.....)))))........)))))))))).))-------).......(((........))).-- ( -20.59, z-score = -2.10, R) >dp4.chrXR_group6 12383808 80 + 13314419 ----UAUACGUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCAGACAAACAGACGUAACGUG-------CAAAAAGGAACAAAACACGA----- ----..(((((.((((((((........(((((.....)))))........)))))))))))))-------....................----- ( -19.59, z-score = -2.32, R) >droPer1.super_12 803967 80 + 2414086 ----UAUACGUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCAGACAAACAGACGUAACGUG-------CAAAAAGGAACAAAACACGA----- ----..(((((.((((((((........(((((.....)))))........)))))))))))))-------....................----- ( -19.59, z-score = -2.32, R) >consensus ____UAUGGCUAUAUGUCUGGCAUAUAAACAGUGCUCAGCUGUCUGACAAACAGACGUAACGUGC______AAAAAAGGAGGAAAACACAGUAGGG ............((((((((........(((((.....)))))........))))))))..................................... (-14.99 = -14.99 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:41 2011