| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,792,796 – 14,792,888 |
| Length | 92 |
| Max. P | 0.928787 |

| Location | 14,792,796 – 14,792,888 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 68.48 |
| Shannon entropy | 0.60685 |
| G+C content | 0.58111 |
| Mean single sequence MFE | -27.11 |
| Consensus MFE | -15.91 |
| Energy contribution | -16.03 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14792796 92 + 24543557 ---UCUGCCA--CGCCCCCAAGGGCGCCUCAGGCGCCGCCCAUGUGGCU-CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAAUGAAAAUCGAA- ---(((((..--(((((....))))).....((.(((((....))))).-))..)).......(((((((.....)))))))----))).............- ( -34.30, z-score = -1.50, R) >droMoj3.scaffold_6680 22994318 72 + 24764193 ------------------UAUGCACGCCCUUGGCGCUGUCAUUUCGUAU--GUCGCA------GGGUCGCUCUGAGCACCC-----AGCCCUUAAAAACUAAA ------------------..(((.((((...))))..(.(((.....))--).))))------((((.((.....))))))-----................. ( -17.10, z-score = -0.07, R) >droVir3.scaffold_13049 17893721 71 + 25233164 ------------------CACGCACGCCCUUGGCGCUGUCAUUUCGCAU--GUAGCA------GGGUAGCUCUGAGCACCC-----AGCCCUUGAAA-CUAAA ------------------.............(((((((.(((.....))--))))).------((((.((.....))))))-----.))).......-..... ( -18.20, z-score = 0.48, R) >droPer1.super_12 1632315 100 + 2414086 GAUCCCGAUAGUCCCCUCCAAGUACGCCACAGGCGCCAUUUAU-CGGCU-CGCAGCAUCCAAAGGGUUGCUCUGAGCAGCCUUACAAAAAACAAAAAACAAA- .....((((((.............((((...))))....))))-))(((-((.((((.((....)).)))).))))).........................- ( -21.73, z-score = -1.04, R) >dp4.chrXR_group6 952694 100 + 13314419 GAUCCCGAUAGUCCCCUCCAAGUACGCCACAGGCGCCAUUUAU-CGGCU-CGCAGCAUCCAAAGGGUUGCUCUGAGCAGCCUUACAAAAAACAAAAAACAAA- .....((((((.............((((...))))....))))-))(((-((.((((.((....)).)))).))))).........................- ( -21.73, z-score = -1.04, R) >droEre2.scaffold_4784 14760397 92 + 25762168 ---UCUGCCA--CGCCCCCAAGGCCGCCUCAGGCGCCGCCCACGUGGCU-CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAAUGAAAACCUAA- ---(((((((--((.......(((.(((...)))))).....)))))).-.............(((((((.....)))))))----))).............- ( -32.30, z-score = -1.81, R) >droYak2.chr3L 14881285 92 + 24197627 ---UCUGCCA--CGCCCCCAAGGCCGCCAGAGGCGUCGCCCACGUGGCU-CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAAUGAAAACCCAA- ---(((((((--((((.....)))((((...))))........))))).-.............(((((((.....)))))))----))).............- ( -31.10, z-score = -1.42, R) >droSec1.super_0 6913937 93 + 21120651 ---UCUGCCA--CGCCCCCAAGGGCGCCUCAGGCGCCGCCCAUGUGGCUUCCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAAUGAAAAUCUAA- ---(((((((--(((((....))))(((...))).........)))))...............(((((((.....)))))))----))).............- ( -33.70, z-score = -1.53, R) >droSim1.chr3L 14123205 93 + 22553184 ---UCUGCCA--CGCCCCCAAGGGCGCCUCAGGCGCCGCCCAUGUGGCUUCCCAGCAUCCUACGGGUUGCUCUGAGCAGCCC----AGAAAUGAAAAUCUAA- ---.((((..--(((((....)))))..((((..(((((....))))).....((((.((....)).))))))))))))..(----(....)).........- ( -33.80, z-score = -1.53, R) >consensus ___UCUGCCA__CGCCCCCAAGGACGCCUCAGGCGCCGCCCAUGUGGCU_CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC____AGAAAUGAAAAACUAA_ .....................((..(((...)))((((......))))...))..........(((((((.....)))))))..................... (-15.91 = -16.03 + 0.13)

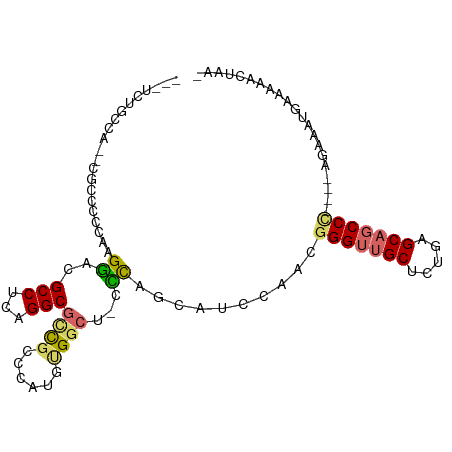
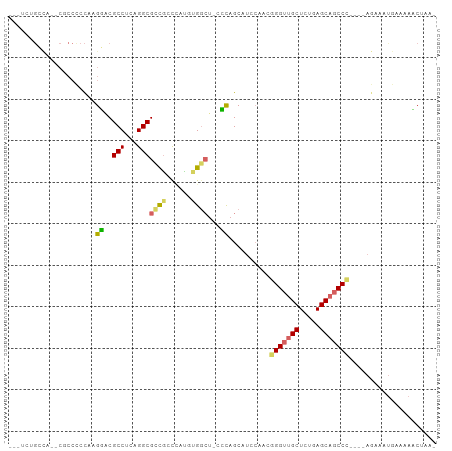
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:39 2011