
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,245,678 – 5,245,777 |
| Length | 99 |
| Max. P | 0.979409 |

| Location | 5,245,678 – 5,245,769 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 89.22 |
| Shannon entropy | 0.21165 |
| G+C content | 0.44024 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -21.92 |
| Energy contribution | -22.25 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
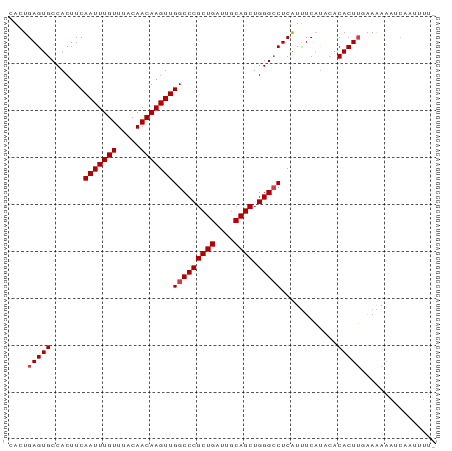
>dm3.chr2L 5245678 91 + 23011544 CACUGAGUGCCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUACACACUUUAAAAAAUCAAUUUUC ....(((((.......(((((((.....)))))))(((((((((....)))).)))))............)))))................ ( -23.50, z-score = -2.89, R) >droSim1.chr2L 5043967 91 + 22036055 CACUGAGUGCCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUGCACACUUGGAAAAAUCAAUUUUU ..(..((((.......(((((((.....)))))))(((((((((....)))).)))))............))))..).............. ( -27.50, z-score = -2.65, R) >droSec1.super_5 3325972 90 + 5866729 CACUGAGUGCCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUGCACACUUGGAAAAAUCAAUUUU- ..(..((((.......(((((((.....)))))))(((((((((....)))).)))))............))))..).............- ( -27.50, z-score = -2.56, R) >droYak2.chr2L 8371467 91 - 22324452 CACUGAGUGCCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUCCACACUUCAAAAAAUCUAUGUUU ....(((((.......(((((((.....)))))))(((((((((....)))).)))))............)))))................ ( -23.50, z-score = -2.30, R) >droEre2.scaffold_4929 5323545 91 + 26641161 CACUGAGUGCCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUCCACACUUCAAGAAAUCAAUGUUG ..(((((((.......(((((((.....)))))))(((((((((....)))).)))))............)))))..))............ ( -23.60, z-score = -1.64, R) >droAna3.scaffold_12916 6261915 81 - 16180835 CACUGAGUGCCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGACUCGUCUCCCAAUCACUGGAAGAA---------- ....((((.(((..(((((((((.....)))))))))...((((....))))))).)))).(((.(((.....))).))).---------- ( -25.90, z-score = -2.15, R) >consensus CACUGAGUGCCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUACACACUUGAAAAAAUCAAUUUU_ ....(((((.......(((((((.....)))))))(((((((((....)))).)))))............)))))................ (-21.92 = -22.25 + 0.33)


| Location | 5,245,687 – 5,245,777 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.66 |
| Shannon entropy | 0.36831 |
| G+C content | 0.43413 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
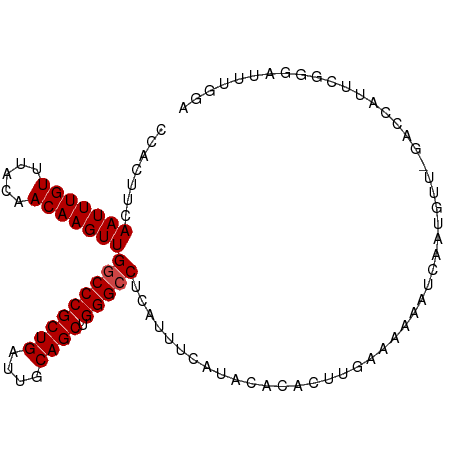
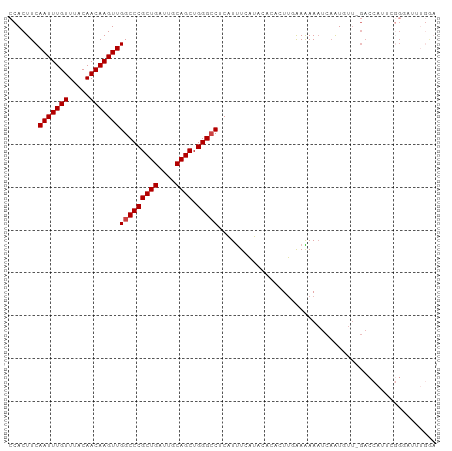
>dm3.chr2L 5245687 90 + 23011544 CCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUACACACUUUAAAAAAUCAAUUUUCGACUUGCA---------- .((..(((((((((.....)))))))(((((((((....)))).))))).................................))..))..---------- ( -20.10, z-score = -2.38, R) >droSim1.chr2L 5043976 100 + 22036055 CCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUGCACACUUGGAAAAAUCAAUUUUUGACCAUUCGGUAUUUGGA (((....(((((((.....)))))))(((((((((....)))).))))).......((((.....(((.((((.....))))..)))....)))).))). ( -25.90, z-score = -1.42, R) >droSec1.super_5 3325981 99 + 5866729 CCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUGCACACUUGGAAAAAUCAAUUUU-GACCAUUCGGGAUUUGAA (((....(((((((.....)))))))(((((((((....)))).)))))................))).....((((((((-((....)))))).)))). ( -25.80, z-score = -1.42, R) >droYak2.chr2L 8371476 100 - 22324452 CCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUCCACACUUCAAAAAAUCUAUGUUUGACCAUUCGGGAUUUGGA (((..(((((((((.....)))))))(((((((((....)))).))))).........((.....(((((.........)))))......))))..))). ( -25.00, z-score = -1.40, R) >droEre2.scaffold_4929 5323554 100 + 26641161 CCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUCCACACUUCAAGAAAUCAAUGUUGGCCCAUUAGGGAUUUGGA (((..(((((((((.....)))))))))(((((((....))))((((((.((((.....................))))..))))))...)))...))). ( -30.00, z-score = -1.86, R) >droAna3.scaffold_12916 6261924 77 - 16180835 CCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGACUCGUCUCCCAAUCACUGGAAGAA--UAGUG--------------------- .....(((((((((.....)))))))))(((((((....)))).)))(((..(((.(((.....))).))).--.))).--------------------- ( -22.80, z-score = -2.13, R) >consensus CCACUUCAAUUUGUUUACAACAAGUUGGCCCGCUGAUUGCAGCUGGGCCUCAUUUCAUACACACUUGAAAAAAUCAAUGUU_GACCAUUCGGGAUUUGGA .......(((((((.....)))))))(((((((((....)))).)))))................................................... (-18.42 = -18.58 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:27 2011