| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,743,443 – 14,743,535 |
| Length | 92 |
| Max. P | 0.999853 |

| Location | 14,743,443 – 14,743,535 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 68.44 |
| Shannon entropy | 0.60444 |
| G+C content | 0.32092 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -4.49 |
| Energy contribution | -5.10 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
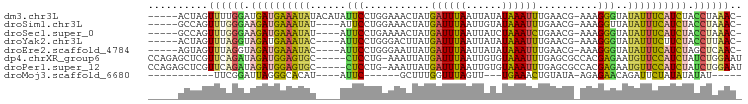
>dm3.chr3L 14743443 92 + 24543557 -GUUUAGGUAGAUGAAAUAUACCCUUU-CGUUCAAAUUUAUAUAAUUAAAUCAUAGUUUCCAGGAAUAUGUAUAUUUCAUCAUCCAAAACUAGU----- -((((.((..((((((((((((...((-(.(..(((((................)))))..).)))...))))))))))))..)).))))....----- ( -19.79, z-score = -3.98, R) >droSim1.chr3L 14073769 88 + 22553184 -GUUUAGGUAGAUGAAAUAUAACCUUU-CGUUCAAAUUUAUACAAUUAAAUCAUAGUUUCCAGGAAU----AUAUUUCAUCUUCCCAAACUGGC----- -((((.((.((((((((((((.(((..-.......(((((......)))))..........)))..)----))))))))))).)).))))....----- ( -20.01, z-score = -3.85, R) >droSec1.super_0 6864433 88 + 21120651 -GUUUAGGUAGAUGAAAUAUACCCUUU-CGUUCAGAUUUAGAUAAUUAAAUCAUAGUUUUCAGGAAU----AUAUUUCAUCUUCCCAAACUGGC----- -((((.((.((((((((((((.(((..-......(((((((....))))))).........)))..)----))))))))))).)).))))....----- ( -22.43, z-score = -3.89, R) >droYak2.chr3L 14830567 88 + 24197627 -GUUAAGGUAGAAGAAAUAUACCCUUU-CGUUCAAAUUUAUAUAAUUAAAUCAUAAGUCCCAGGAAU----GUAUUUCAUCUACCUAAACUAGU----- -(((.(((((((.((((((((.(((..-.......(((((......)))))..........)))..)----))))))).))))))).)))....----- ( -21.01, z-score = -4.50, R) >droEre2.scaffold_4784 14709917 88 + 25762168 -GUUGAGCUAGAUGAAAUAUACCCUUU-CGUUCAAAUUUAUAUAAUUAAAUCAUAAUUCCCAGGAAU----GUAUUUCAUCUACCUAAACUACU----- -(((.((.(((((((((((((.(((..-.......(((((......)))))..........)))..)----)))))))))))).)).)))....----- ( -18.71, z-score = -4.41, R) >dp4.chrXR_group6 12308461 93 - 13314419 AUUCCAGAUAGAUGGAACAUUCUCGUGGCGCUCAAAUUUACACAAUUAAAUCAUAAUUU-CAGGAG-----GCACUCCAUCUAUCUGAACGAGCUCUGG ....((((((((((((....((((.(((.......(((((......))))).......)-)).)))-----)...))))))))))))............ ( -25.04, z-score = -3.22, R) >droPer1.super_12 729395 93 - 2414086 AUUCCAGAUAGAUGGAACAUUCUCGUGGCGCUCAAAUUUACACAAUUAAAUCAUAAUUU-CAGGAG-----GCACUCCAUCUAUCUGAACGAGCUCUGG ....((((((((((((....((((.(((.......(((((......))))).......)-)).)))-----)...))))))))))))............ ( -25.04, z-score = -3.22, R) >droMoj3.scaffold_6680 4911329 69 - 24764193 -----AUAUAUAUAGAAUCUGUUCUCU-UAUACAGUUUCA---AACUAAACCAAAGC------GAAU----AUGUGCCCUAAUCCGAA----------- -----.(((((((.(((.((((.....-...)))).))).---..((.......)).------..))----)))))............----------- ( -6.50, z-score = -0.72, R) >consensus _GUUUAGGUAGAUGAAAUAUACCCUUU_CGUUCAAAUUUAUAUAAUUAAAUCAUAAUUUCCAGGAAU____AUAUUUCAUCUACCUAAACUAGC_____ ......((.(((((((((.......................................................))))))))).)).............. ( -4.49 = -5.10 + 0.61)

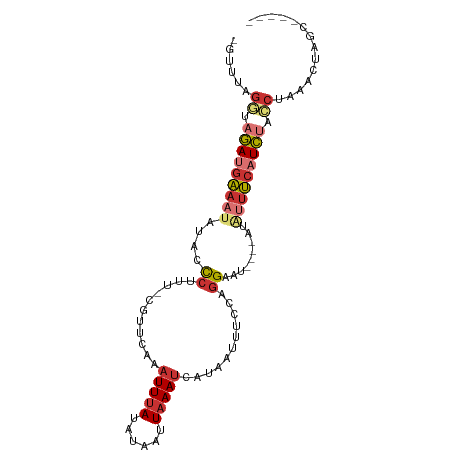
| Location | 14,743,443 – 14,743,535 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 68.44 |
| Shannon entropy | 0.60444 |
| G+C content | 0.32092 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -11.28 |
| Energy contribution | -10.55 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.64 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14743443 92 - 24543557 -----ACUAGUUUUGGAUGAUGAAAUAUACAUAUUCCUGGAAACUAUGAUUUAAUUAUAUAAAUUUGAACG-AAAGGGUAUAUUUCAUCUACCUAAAC- -----....((((.((..((((((((((((........(....).........................(.-...).))))))))))))..)).))))- ( -21.30, z-score = -3.56, R) >droSim1.chr3L 14073769 88 - 22553184 -----GCCAGUUUGGGAAGAUGAAAUAU----AUUCCUGGAAACUAUGAUUUAAUUGUAUAAAUUUGAACG-AAAGGUUAUAUUUCAUCUACCUAAAC- -----....(((((((.(((((((((((----(..(((.........((((((......))))))......-..))).)))))))))))).)))))))- ( -26.83, z-score = -5.30, R) >droSec1.super_0 6864433 88 - 21120651 -----GCCAGUUUGGGAAGAUGAAAUAU----AUUCCUGAAAACUAUGAUUUAAUUAUCUAAAUCUGAACG-AAAGGGUAUAUUUCAUCUACCUAAAC- -----....(((((((.(((((((((((----((.(((.........((((((......))))))......-..)))))))))))))))).)))))))- ( -29.93, z-score = -6.62, R) >droYak2.chr3L 14830567 88 - 24197627 -----ACUAGUUUAGGUAGAUGAAAUAC----AUUCCUGGGACUUAUGAUUUAAUUAUAUAAAUUUGAACG-AAAGGGUAUAUUUCUUCUACCUUAAC- -----....(((.(((((((.((((((.----..((((.........((((((......))))))......-..))))..)))))).))))))).)))- ( -21.53, z-score = -2.96, R) >droEre2.scaffold_4784 14709917 88 - 25762168 -----AGUAGUUUAGGUAGAUGAAAUAC----AUUCCUGGGAAUUAUGAUUUAAUUAUAUAAAUUUGAACG-AAAGGGUAUAUUUCAUCUAGCUCAAC- -----....(((.((.(((((((((((.----..((((.........((((((......))))))......-..))))..))))))))))).)).)))- ( -21.43, z-score = -3.18, R) >dp4.chrXR_group6 12308461 93 + 13314419 CCAGAGCUCGUUCAGAUAGAUGGAGUGC-----CUCCUG-AAAUUAUGAUUUAAUUGUGUAAAUUUGAGCGCCACGAGAAUGUUCCAUCUAUCUGGAAU ..........(((((((((((((((...-----(((.((-.(((((.....)))))((((........)))))).)))....))))))))))))))).. ( -27.50, z-score = -3.12, R) >droPer1.super_12 729395 93 + 2414086 CCAGAGCUCGUUCAGAUAGAUGGAGUGC-----CUCCUG-AAAUUAUGAUUUAAUUGUGUAAAUUUGAGCGCCACGAGAAUGUUCCAUCUAUCUGGAAU ..........(((((((((((((((...-----(((.((-.(((((.....)))))((((........)))))).)))....))))))))))))))).. ( -27.50, z-score = -3.12, R) >droMoj3.scaffold_6680 4911329 69 + 24764193 -----------UUCGGAUUAGGGCACAU----AUUC------GCUUUGGUUUAGUU---UGAAACUGUAUA-AGAGAACAGAUUCUAUAUAUAU----- -----------...((((((((((....----....------))))))))))....---.(((.((((...-.....)))).))).........----- ( -12.40, z-score = -1.23, R) >consensus _____GCUAGUUUAGGAAGAUGAAAUAC____AUUCCUGGAAACUAUGAUUUAAUUAUAUAAAUUUGAACG_AAAGGGUAUAUUUCAUCUACCUAAAC_ ..........((((((.((((((((((......(((...........((((((......))))))..........)))..)))))))))).)))))).. (-11.28 = -10.55 + -0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:33 2011