| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,710,241 – 14,710,338 |
| Length | 97 |
| Max. P | 0.735281 |

| Location | 14,710,241 – 14,710,338 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 68.25 |
| Shannon entropy | 0.64464 |
| G+C content | 0.34618 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -6.45 |
| Energy contribution | -7.38 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
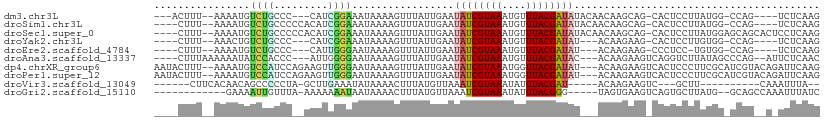
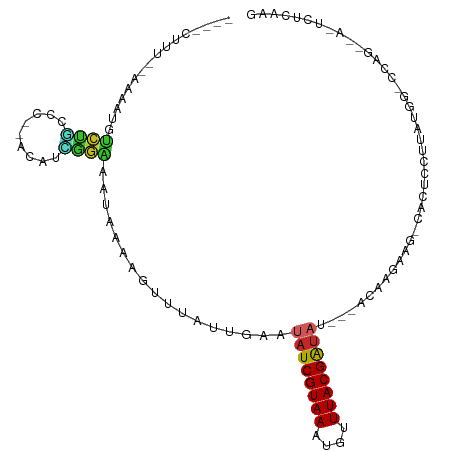
>dm3.chr3L 14710241 97 + 24543557 CUUGAGA----CUGG-CCAUAAGGAGUG-CUGCUUGUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUG---GGGCAGACAUUUU--AAAGU--- .((((((----(((.-((....((((..-..(.((((((.(((((((((....))))))))).)))))).)......))))....---)).)))...))))--))...--- ( -26.30, z-score = -2.90, R) >droSim1.chr3L 14042986 99 + 22553184 CUUGAGA----CUGG-CCAUAAGGAGUG-CUGCUUGUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUGUGGGGGCAGACAUUUU--AAAG---- .......----....-......((((((-((((((((((.(((((((((....))))))))).)))))).........(((......))))))).))))))--....---- ( -24.70, z-score = -2.04, R) >droSec1.super_0 6835313 104 + 21120651 CUUGAGGAGUGCUGCUCCAUAAGGAGUG-CUGCUUGUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUGUGGGGGCAGACAUUUU--AAAG---- .....((((((((((((((((.((((..-..(.((((((.(((((((((....))))))))).)))))).)......))))..))).))))))).))))))--....---- ( -34.40, z-score = -4.30, R) >droYak2.chr3L 14800949 93 + 24197627 CUUGAGA----CUGG-CCACAAGGAGUG-CUUCUUGU---AUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUG---GGGCAGACAGUUU--AAAG---- (((.(((----((((-(((((((((...-.)))))))---(((((((((....))))))))).......................---.)))...))))))--.)))---- ( -23.60, z-score = -2.67, R) >droEre2.scaffold_4784 14681107 92 + 25762168 CUUGAGA----CUGG-CCACA-GGAGGG-CUUCUUGU---AUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUCCCAAUG---GGGCAGACAUUUU--AAAG---- .((((((----(((.-((.((-((((((-.((.(((.---(((((((((....))))))))).))).)).))....))))...))---)).)))...))))--))..---- ( -20.50, z-score = -1.84, R) >droAna3.scaffold_13337 18906544 99 - 23293914 GUUGAGAAU--CUGGGCUAUAAGACCUGACUUCUUGU---GUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUCCCCAAU---GGGUGGAUAUUUUUUAAAG---- (((((((((--(.((.........)).)).)))))(.---(((((((((....))))))))).)....)))...((((((((...---))).)))))..........---- ( -20.10, z-score = -1.19, R) >dp4.chrXR_group6 12281669 106 - 13314419 CUUGAAUCUGUACGAUGCGAAGGGAGUGACUUCUUGU---AUAUCGUAACCAUUUACGAUAUUCAAUAAACUUUUAUUCCCAACUUCUGGAUGGACAUUUU--AAAGUAUU .(((.(((.....))).))).((((((((.((.(((.---(((((((((....))))))))).))).))....)))))))).((((..((((....)))).--.))))... ( -22.90, z-score = -1.70, R) >droPer1.super_12 702349 106 - 2414086 CUUGAAUCUGUACGAUGCGAAGGGAGUGACUUCUUGU---AUAUCGUAACCAUUUACGAUAUUCAAUAAACUUUUAUUCCCAACUUCUGGAUGGACAUUUU--AAAGUAUU .(((.(((.....))).))).((((((((.((.(((.---(((((((((....))))))))).))).))....)))))))).((((..((((....)))).--.))))... ( -22.90, z-score = -1.70, R) >droVir3.scaffold_13049 8421955 84 - 25233164 --UAAAUUUG----------AAGC---GACUUCUUGU-----AUCGUAAAUAUUUACGAUUUAACAUAAAGUUUUAUAUUUCAAGC-UAGGGGGCUGUUGUGAAG------ --........----------....---(((((..(((-----(((((((....)))))))...)))..)))))(((((.....(((-(....))))..)))))..------ ( -14.80, z-score = -0.75, R) >droGri2.scaffold_15110 16297941 91 + 24565398 GAUAAAUUUGGCUGC--CAUAAGCACUGACUUCACUA-----CCCGUAAAUAUUUACGAUUUAACAUAAAGUUUUAUUAUUUUUUU-UAAACAAUUUUC------------ ((((((....(((..--....))).............-----..(((((....)))))..............))))))........-............------------ ( -7.50, z-score = 0.04, R) >consensus CUUGAGA_U__CUGG_CCAUAAGGAGUG_CUUCUUGU___AUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUGU__GGGCAGACAUUUU__AAAG____ .................................((((...(((((((((....)))))))))...)))).......................................... ( -6.45 = -7.38 + 0.93)


| Location | 14,710,241 – 14,710,338 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 68.25 |
| Shannon entropy | 0.64464 |
| G+C content | 0.34618 |
| Mean single sequence MFE | -18.07 |
| Consensus MFE | -5.84 |
| Energy contribution | -6.05 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14710241 97 - 24543557 ---ACUUU--AAAAUGUCUGCCC---CAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAACAAGCAG-CACUCCUUAUGG-CCAG----UCUCAAG ---.....--.....(.(((..(---(((.(((.......((((.(((.(((((((((....))))))))).))).))))..-...)))..))))-.)))----.)..... ( -20.00, z-score = -2.60, R) >droSim1.chr3L 14042986 99 - 22553184 ----CUUU--AAAAUGUCUGCCCCCACAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAACAAGCAG-CACUCCUUAUGG-CCAG----UCUCAAG ----....--.....(.(((((.((.....))........((((.(((.(((((((((....))))))))).))).))))..-..........))-.)))----.)..... ( -19.30, z-score = -2.37, R) >droSec1.super_0 6835313 104 - 21120651 ----CUUU--AAAAUGUCUGCCCCCACAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAACAAGCAG-CACUCCUUAUGGAGCAGCACUCCUCAAG ----....--....................(((.......((((.(((.(((((((((....))))))))).))).)))).(-(.((((....))))..))..)))..... ( -23.90, z-score = -3.39, R) >droYak2.chr3L 14800949 93 - 24197627 ----CUUU--AAACUGUCUGCCC---CAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAU---ACAAGAAG-CACUCCUUGUGG-CCAG----UCUCAAG ----....--..((((...(((.---((..(((.......((((.(((.(((((((((....)))))))))---.))).)))-)..))).)).))-))))----)...... ( -20.60, z-score = -2.64, R) >droEre2.scaffold_4784 14681107 92 - 25762168 ----CUUU--AAAAUGUCUGCCC---CAUUGGGAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAU---ACAAGAAG-CCCUCC-UGUGG-CCAG----UCUCAAG ----....--.....(.(((((.---((..(((.......((((.(((.(((((((((....)))))))))---.))).)))-))))..-)).))-.)))----.)..... ( -20.81, z-score = -2.48, R) >droAna3.scaffold_13337 18906544 99 + 23293914 ----CUUUAAAAAAUAUCCACCC---AUUGGGGAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAC---ACAAGAAGUCAGGUCUUAUAGCCCAG--AUUCUCAAC ----............(((.((.---...)))))...........(((..((((((((....)))))))).---.)))((((((.(((......)))..)--))).))... ( -16.60, z-score = -0.77, R) >dp4.chrXR_group6 12281669 106 + 13314419 AAUACUUU--AAAAUGUCCAUCCAGAAGUUGGGAAUAAAAGUUUAUUGAAUAUCGUAAAUGGUUACGAUAU---ACAAGAAGUCACUCCCUUCGCAUCGUACAGAUUCAAG ....((.(--(..((((...(((........)))...........(((.(((((((((....)))))))))---.)))((((.......))))))))..)).))....... ( -17.20, z-score = -0.45, R) >droPer1.super_12 702349 106 + 2414086 AAUACUUU--AAAAUGUCCAUCCAGAAGUUGGGAAUAAAAGUUUAUUGAAUAUCGUAAAUGGUUACGAUAU---ACAAGAAGUCACUCCCUUCGCAUCGUACAGAUUCAAG ....((.(--(..((((...(((........)))...........(((.(((((((((....)))))))))---.)))((((.......))))))))..)).))....... ( -17.20, z-score = -0.45, R) >droVir3.scaffold_13049 8421955 84 + 25233164 ------CUUCACAACAGCCCCCUA-GCUUGAAAUAUAAAACUUUAUGUUAAAUCGUAAAUAUUUACGAU-----ACAAGAAGUC---GCUU----------CAAAUUUA-- ------.........(((..(...-.((((.(((((((....)))))))..(((((((....)))))))-----.))))..)..---))).----------........-- ( -12.60, z-score = -2.48, R) >droGri2.scaffold_15110 16297941 91 - 24565398 ------------GAAAAUUGUUUA-AAAAAAAUAAUAAAACUUUAUGUUAAAUCGUAAAUAUUUACGGG-----UAGUGAAGUCAGUGCUUAUG--GCAGCCAAAUUUAUC ------------....(((((((.-....)))))))...(((((((......((((((....)))))).-----..)))))))...(((.....--)))............ ( -12.50, z-score = -0.51, R) >consensus ____CUUU__AAAAUGUCUGCCC__ACAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAU___ACAAGAAG_CACUCCUUAUGG_CCAG__A_UCUCAAG ..................................................((((((((....))))))))......................................... ( -5.84 = -6.05 + 0.21)
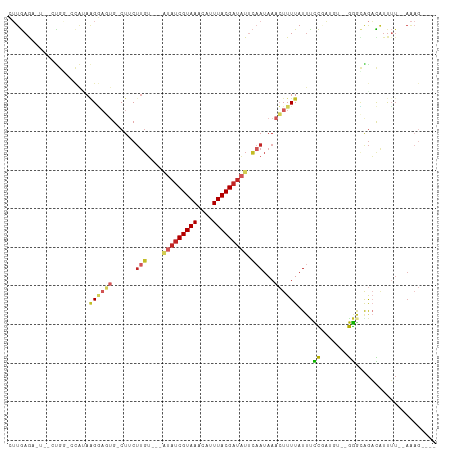
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:29 2011