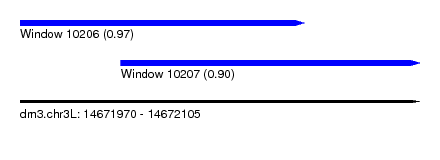
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,671,970 – 14,672,105 |
| Length | 135 |
| Max. P | 0.974179 |

| Location | 14,671,970 – 14,672,066 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.64 |
| Shannon entropy | 0.33708 |
| G+C content | 0.28365 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -13.02 |
| Energy contribution | -14.65 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14671970 96 + 24543557 AUAACCGUUAGUCUUACAAGGUUUCAAUAAAGAAAUAAGAAAUGUGGCAAUGACGCAUUAUACUUAUAUAAUGGGUCAUUAAAGCCACGUUUUUUA------------ ..((((.............))))............(((((((((((((((((((.(((((((....))))))).))))))...)))))))))))))------------ ( -30.12, z-score = -5.76, R) >droEre2.scaffold_4784 14642353 107 + 25762168 AUAACCGUUAGUCUUACAAAGUUUCAAUAAAGCAAUAAGAUAGGUAG-AAUGACCCAUUAUACUUAUAUAAUAGGUCAAUUAAUCGUCCAUUCAAUCAGUUCAGUUCA ...(((....((((((....((((.....))))..)))))).)))..-..(((((.((((((....)))))).))))).............................. ( -15.80, z-score = -2.25, R) >droSec1.super_0 6797088 96 + 21120651 AUAACCGUUAGUAUUACAAGGUUUCAAUAAAGAAAUAAGAAAUGUGGCAAUGACGCGUUAUACUUAUAUAAUGGAUCAUUAAAGCAACAUUUUAUA------------ ....................(((((......)))))..(((((((.(((((((..(((((((....)))))))..)))))...)).)))))))...------------ ( -16.80, z-score = -1.52, R) >droSim1.chr3L 14003561 99 + 22553184 AUAACUGUUAGUUUUACAAAGUUUCAAUAAAGAAAAAAGAAAUGUGGCAAUGACGCGUUAUACUUAUGUAAUGGUUCAUAAAAGCAACGUUUUUUUUUA--------- .................................((((((((((((.((.(((((.(((((((....)))))))).))))....)).)))))))))))).--------- ( -16.50, z-score = -0.44, R) >consensus AUAACCGUUAGUCUUACAAAGUUUCAAUAAAGAAAUAAGAAAUGUGGCAAUGACGCAUUAUACUUAUAUAAUGGGUCAUUAAAGCAACGUUUUAUA____________ ....................(((((......)))))..(((((((.((((((((.(((((((....))))))).))))))...)).)))))))............... (-13.02 = -14.65 + 1.63)


| Location | 14,672,004 – 14,672,105 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Shannon entropy | 0.38728 |
| G+C content | 0.29183 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.83 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14672004 101 + 24543557 AUAAGAAAUGUGGCAAUGACGCAUUAUACUUAUAUAAUGGGUCAUUAAAGCCACGUUUUUU---AUUCAGU-------AUCGCUUUU---AAUAGUUUUGGUGCACUUCAGUUA ((((((((((((((((((((.(((((((....))))))).))))))...))))))))))))---))...((-------((((((...---...)))...))))).......... ( -34.00, z-score = -6.07, R) >droEre2.scaffold_4784 14642387 113 + 25762168 AUAAGAUAGGUAG-AAUGACCCAUUAUACUUAUAUAAUAGGUCAAUUAAUCGUCCAUUCAAUCAGUUCAGUUCAUUCAAUCGUUUUUUGUAAUAGUUUUACAGCACUUUAGGUU ....(((.(((.(-(.(((((.((((((....)))))).))))).)).)))))).......................((((.....((((((.....)))))).......)))) ( -15.20, z-score = -0.89, R) >droSec1.super_0 6797122 100 + 21120651 AUAAGAAAUGUGGCAAUGACGCGUUAUACUUAUAUAAUGGAUCAUUAAAGCAACAUUUUAU---AUUCAAU-------AUUGCUUUU---AAUAGUUUUAACGCACUUCAGUU- ....(((.((((..(((((..(((((((....)))))))..)))))(((((((.(((....---....)))-------.))))))).---...........)))).)))....- ( -19.00, z-score = -1.65, R) >droSim1.chr3L 14003595 104 + 22553184 AAAAGAAAUGUGGCAAUGACGCGUUAUACUUAUGUAAUGGUUCAUAAAAGCAACGUUUUUUUUUAUUGAAU-------AUAGCUUUU---AAUAGUUUUAGCGCACUUGAGUUA (((((((((((.((.(((((.(((((((....)))))))).))))....)).)))))))))))........-------.((((((..---....((....))......)))))) ( -18.20, z-score = 0.38, R) >consensus AUAAGAAAUGUGGCAAUGACGCAUUAUACUUAUAUAAUGGGUCAUUAAAGCAACGUUUUAU___AUUCAAU_______AUCGCUUUU___AAUAGUUUUAGCGCACUUCAGUUA ....(((((((.((((((((.(((((((....))))))).))))))...)).)))))))....................................................... (-12.70 = -13.83 + 1.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:25 2011