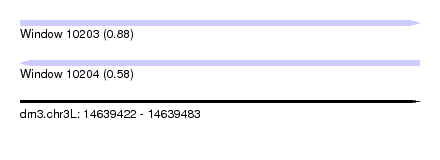
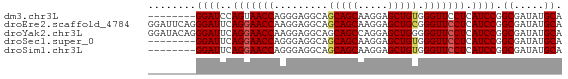
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,639,422 – 14,639,483 |
| Length | 61 |
| Max. P | 0.878672 |

| Location | 14,639,422 – 14,639,483 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Shannon entropy | 0.18370 |
| G+C content | 0.58888 |
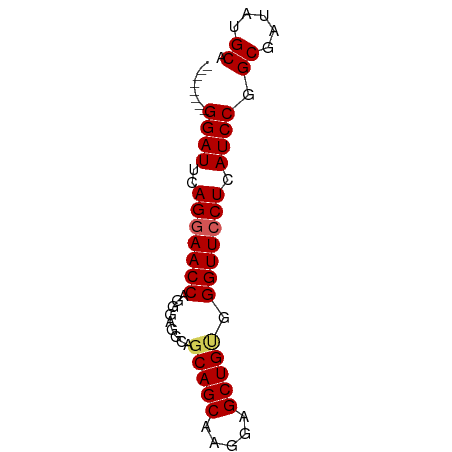
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
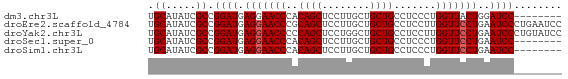
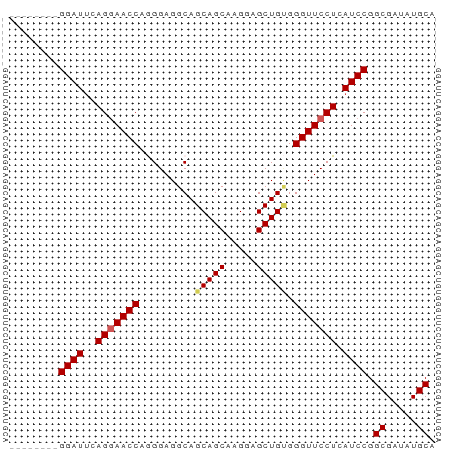
>dm3.chr3L 14639422 61 + 24543557 UGCAUAUCGCCGGAUGAGGAACCCACAGCUCCUUGCUGCUGCCUCCCUGGUUACUGGAUCC-------- ........(((((..((((......((((.....))))...)))).)))))..........-------- ( -16.00, z-score = -0.34, R) >droEre2.scaffold_4784 14609604 69 + 25762168 UGCAUAUCGCCGGAUGAGGAACCCGCAGCUCCUUGCUGCUGCCUCCUUGGUUCCUGAAUCCCUGAAUCC .((.....)).((((.(((((((.(((((........)))))......)))))))..))))........ ( -22.60, z-score = -2.94, R) >droYak2.chr3L 14728007 69 + 24197627 UGCAUAUCGCCGGAUGAGGAACCCCCAGCUCCUGGCUGCUGCCUCCUUGGUUCCUGAAUCCCUGUAUCC .((.....)).((((.(((((((..((((........)))).......)))))))..))))........ ( -19.00, z-score = -1.13, R) >droSec1.super_0 6764759 61 + 21120651 UGCAUAUCGCCGGAUGAGGAACCCACAGCUCCUUGCUGCUGCCUCCCUGGUUCCUGAAUCC-------- .((.....)).((((.(((((((((((((.....)))).)).......)))))))..))))-------- ( -19.01, z-score = -2.17, R) >droSim1.chr3L 13970026 61 + 22553184 UGCAUAUCGCCGGAUGAGGAACCCACAGCUCCUUGCUGCUGCCUCCCUGGUUCCUGAAUCC-------- .((.....)).((((.(((((((((((((.....)))).)).......)))))))..))))-------- ( -19.01, z-score = -2.17, R) >consensus UGCAUAUCGCCGGAUGAGGAACCCACAGCUCCUUGCUGCUGCCUCCCUGGUUCCUGAAUCC________ .((.....)).((((.(((((((..((((........)))).......)))))))..))))........ (-16.90 = -17.10 + 0.20)
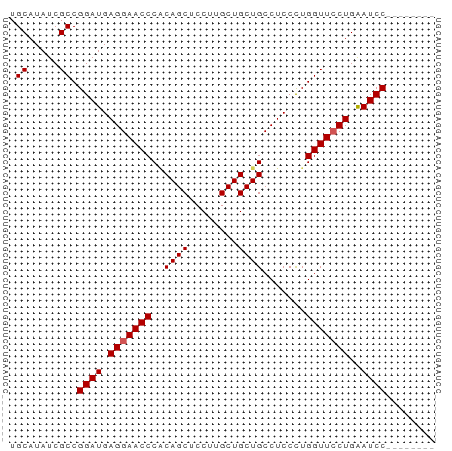
| Location | 14,639,422 – 14,639,483 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Shannon entropy | 0.18370 |
| G+C content | 0.58888 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
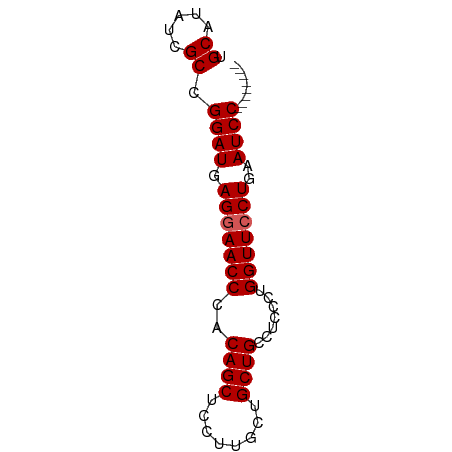
>dm3.chr3L 14639422 61 - 24543557 --------GGAUCCAGUAACCAGGGAGGCAGCAGCAAGGAGCUGUGGGUUCCUCAUCCGGCGAUAUGCA --------((((((........))((((..(((((.....))))).....)))))))).((.....)). ( -17.20, z-score = 0.47, R) >droEre2.scaffold_4784 14609604 69 - 25762168 GGAUUCAGGGAUUCAGGAACCAAGGAGGCAGCAGCAAGGAGCUGCGGGUUCCUCAUCCGGCGAUAUGCA ((((..((((((((.....((.....))..(((((.....))))))))))))).)))).((.....)). ( -23.60, z-score = -1.54, R) >droYak2.chr3L 14728007 69 - 24197627 GGAUACAGGGAUUCAGGAACCAAGGAGGCAGCAGCCAGGAGCUGGGGGUUCCUCAUCCGGCGAUAUGCA ........((((..(((((((..((.(((....))).....))...))))))).)))).((.....)). ( -20.90, z-score = -0.32, R) >droSec1.super_0 6764759 61 - 21120651 --------GGAUUCAGGAACCAGGGAGGCAGCAGCAAGGAGCUGUGGGUUCCUCAUCCGGCGAUAUGCA --------((((..(((((((.........(((((.....))))).))))))).)))).((.....)). ( -21.90, z-score = -1.47, R) >droSim1.chr3L 13970026 61 - 22553184 --------GGAUUCAGGAACCAGGGAGGCAGCAGCAAGGAGCUGUGGGUUCCUCAUCCGGCGAUAUGCA --------((((..(((((((.........(((((.....))))).))))))).)))).((.....)). ( -21.90, z-score = -1.47, R) >consensus ________GGAUUCAGGAACCAGGGAGGCAGCAGCAAGGAGCUGUGGGUUCCUCAUCCGGCGAUAUGCA ........((((..(((((((.........(((((.....))))).))))))).)))).((.....)). (-20.04 = -20.32 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:23 2011