| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,611,020 – 14,611,113 |
| Length | 93 |
| Max. P | 0.969878 |

| Location | 14,611,020 – 14,611,113 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 62.67 |
| Shannon entropy | 0.80636 |
| G+C content | 0.38856 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -9.78 |
| Energy contribution | -9.50 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
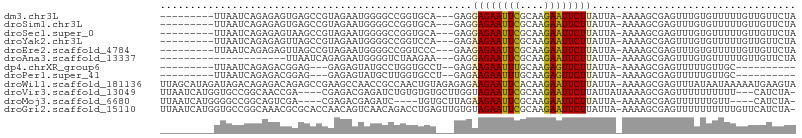
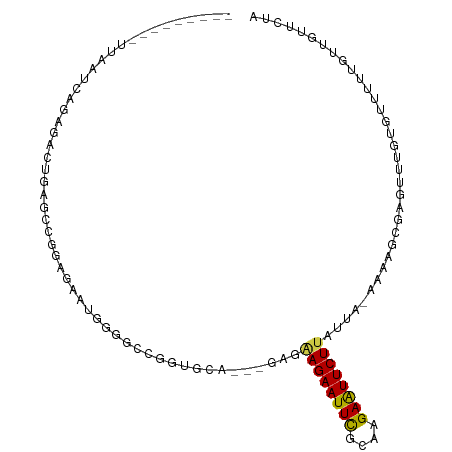
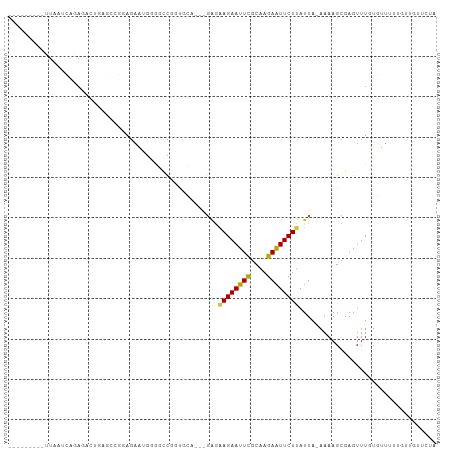
>dm3.chr3L 14611020 93 + 24543557 ---------UUAAUCAGAGAGUGAGCCGUAGAAUGGGGCCGGUGCA---GAGGAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUGUGUUUUUGUUGUUCUA ---------........((((..((((.(.....).))).......---...((((((((....))))))))....-(((((((......))))))).)..)))). ( -21.20, z-score = -0.67, R) >droSim1.chr3L 13938879 93 + 22553184 ---------UUAAUCAGAGAGUGAGCCGUAGAAUGGGGCCGGUGCA---GAGGAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUGUGUUUUUGUUGUUCUA ---------........((((..((((.(.....).))).......---...((((((((....))))))))....-(((((((......))))))).)..)))). ( -21.20, z-score = -0.67, R) >droSec1.super_0 6736510 93 + 21120651 ---------UUAAUCAGAGAGUAAGCCGUAGAAUGGGGCCGGUGCA---GAGGAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUGUGUUUUUGUUGUUCUA ---------...........(((.(((.(.....).)))...)))(---(((((((((((....))))))))....-(((((((......)))))))....)))). ( -21.10, z-score = -0.86, R) >droYak2.chr3L 14699339 93 + 24197627 ---------UUAAUCAGAGAGUUAGCCGUAGAAUGGGGCCGGUCCA---GAGAAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUGUGUUUUUGUUGUUCUA ---------.........(..(..(((.(.....).)))..)..)(---(((((((((((....))))))))....-(((((((......)))))))....)))). ( -21.00, z-score = -1.11, R) >droEre2.scaffold_4784 14580935 93 + 25762168 ---------UUAAUCAGAGAGUUAGCCGUAGAAUGGGGCCGGUCCC---GAAGAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUGUGUUUUUGUUGUUCUA ---------.....((((((...((((((....(((((....))))---)..((((((((....))))))))....-....))).)))....))))))........ ( -23.80, z-score = -1.88, R) >droAna3.scaffold_13337 18809956 81 - 23293914 ---------------------UUAAUCAGAGAAUGGGGUCUAAGAA---GAGGAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUGUGUUUUUGUUGUUCUA ---------------------........((((..(..........---...((((((((....))))))))....-(((((((......))))))).)..)))). ( -18.30, z-score = -1.73, R) >dp4.chrXR_group6 7310064 81 + 13314419 ---------UUAAUCAGAGACGGAG---GAGAGUAUGCCUGGUGCCU--GAGAAGAAUUUGCAAGAGUUCUUAUUA-AAAAGCGAGUUUUUGUUGC---------- ---------.....(((((((..((---(........)))..(((.(--((.((((((((....)))))))).)))-....))).)))))))....---------- ( -14.50, z-score = 0.42, R) >droPer1.super_41 666292 81 + 728894 ---------UUAAUCAGAGACGGAG---GAGAGUAUGCUUGGUGCCU--GAGAAGAAUUUGCAAGAGUUCUUAUUA-AAAAGCGAGUUUUUGUUGC---------- ---------.....(((((((((..---..(((....)))....))(--((.((((((((....)))))))).)))-........)))))))....---------- ( -15.90, z-score = -0.10, R) >droWil1.scaffold_181136 1433700 105 + 2313701 UUAGCAUAGAUAGACAGAGACAGAGCCGAAGCCAACCGCCAACUGUAGAGAGAAGAAUUCACAAGAAUUCUUAUUA-AAAAGCGAGUUUAUAAUAAAAAUGAAGUA .........((((((.......(.((....)))...(((...((......))((((((((....))))))))....-....))).))))))............... ( -14.40, z-score = -1.96, R) >droVir3.scaffold_13049 8532287 98 - 25233164 UUAAUCAUGGUGCCGGCAACCGA----CGAGACGAGAUCUGUGUGUGCUUGGUAGAAUUCGCAAGAAUUCUUAUUAUAAAAGCGAGUUUUUUUUUU---CAUCUA- ........(((.......)))((----.((((.((((....(((.....(((((((((((....)))))).))))).....)))....)))).)))---).))..- ( -21.00, z-score = -0.07, R) >droMoj3.scaffold_6680 20002890 92 + 24764193 UUAAUCAUGGGGCCGGCAGUCGA----CGAGACGAGAUC----UGUGCUUAGAAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUUUUGUU----CAUCUA- ..........(((.....)))((----.(((.(((((..----..(((((..((((((((....))))))))....-..)))))....)))))))----).))..- ( -21.80, z-score = -0.48, R) >droGri2.scaffold_15110 16406146 104 + 24565398 UUAAUCAUGGUGCCGGCAAACGCGCACCAACAGUCAACAGACCUGAGUUGUGUAGAAUUCGCAAGAAUUCUUAUUA-AAAAGCGAGUUUUUUUUUUGUUCAUCUA- .......((((((((.....)).))))))((((..((.((((.((..((..(((((((((....)))))).)))..-.))..)).)))).))..)))).......- ( -26.90, z-score = -2.35, R) >consensus _________UUAAUCAGAGACUGAGCCGGAGAAUGGGGCCGGUGCA___GAGAAGAAUUCGCAAGAAUUCUUAUUA_AAAAGCGAGUUUGUGUUUUUGUUGUUCUA ....................................................((((((((....)))))))).................................. ( -9.78 = -9.50 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:18 2011