| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,596,641 – 14,596,750 |
| Length | 109 |
| Max. P | 0.943444 |

| Location | 14,596,641 – 14,596,750 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.45 |
| Shannon entropy | 0.28655 |
| G+C content | 0.36630 |
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -10.65 |
| Energy contribution | -10.23 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
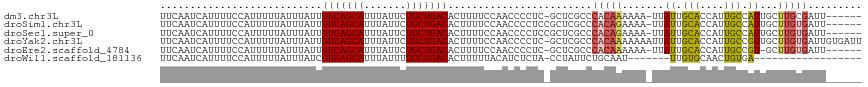
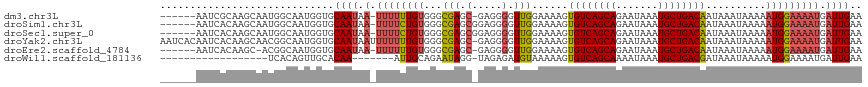
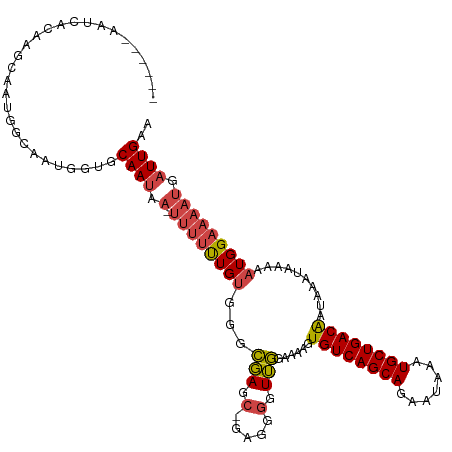
>dm3.chr3L 14596641 109 + 24543557 UUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUC-GCUCGCCCACAAAAAA-UUAUUGCACCAUUGCCAUUGCUUGCGAUU------ ..........................((((((((.......)))))))).............((-((..((..........-..((.(((....))).)).))..))))..------ ( -16.62, z-score = -2.67, R) >droSim1.chr3L 13923943 110 + 22553184 UUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUCCGCUCGCCCACAGAAAA-UUAUUGCACCAUUGCCAUUGCUUGUGAUU------ ..((((.((((((.............((((((((.......))))))))................(....).....)))))-).))))(((....((....))..)))...------ ( -16.30, z-score = -2.25, R) >droSec1.super_0 6721600 110 + 21120651 UUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUCCGCUCGCCCACAGAAAA-UUAUUGCACCAUUGCCAUUGCUUGUGAUU------ ..((((.((((((.............((((((((.......))))))))................(....).....)))))-).))))(((....((....))..)))...------ ( -16.30, z-score = -2.25, R) >droYak2.chr3L 14684788 116 + 24197627 UUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUC-GCUCGCCCACAAAAAAAUUAUUGCACCAUUGCCGUUGCUUGUGAUUGUGAUU ...((((((..((((...........((((((((.......))))))))...............-......................(((.(......).))).)).))..)))))) ( -19.30, z-score = -2.26, R) >droEre2.scaffold_4784 14566150 108 + 25762168 UUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUC-GCUCGCCCACAAAAAA-UUAUUGCACCAUUGCCGU-GCUUGUGAUU------ ...((((((.................((((((((.......))))))))...............-................-.....((((.......))-))..))))))------ ( -17.80, z-score = -2.90, R) >droWil1.scaffold_181136 730594 91 - 2313701 UUCAAUCAUUUUCCAUUUUUAUUUAUCGUCAGCAUUUAUUUUGCUGACACUUUUUACAUCUCUA-CCUAUUCUGCAAU-------UUGUGCAACUGUGA------------------ .....((((..................(((((((.......)))))))................-.......((((..-------...))))...))))------------------ ( -15.10, z-score = -3.28, R) >consensus UUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUC_GCUCGCCCACAAAAAA_UUAUUGCACCAUUGCCAUUGCUUGUGAUU______ ...........................(((((((.......))))))).......................................(((....))).................... (-10.65 = -10.23 + -0.42)
| Location | 14,596,641 – 14,596,750 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Shannon entropy | 0.28655 |
| G+C content | 0.36630 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
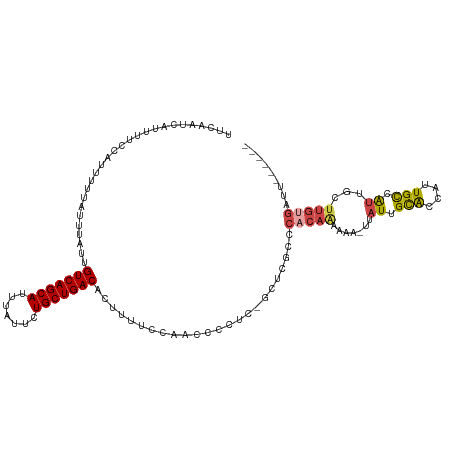
>dm3.chr3L 14596641 109 - 24543557 ------AAUCGCAAGCAAUGGCAAUGGUGCAAUAA-UUUUUUGUGGGCGAGC-GAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAA ------(((((((.((....)).....)))..(((-((((((((......))-)))))))))......((((((((.......))))))))...................))))... ( -23.10, z-score = -1.46, R) >droSim1.chr3L 13923943 110 - 22553184 ------AAUCACAAGCAAUGGCAAUGGUGCAAUAA-UUUUCUGUGGGCGAGCGGAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAA ------...(((..((....))....)))((((.(-((((((((...(.(((......))).).....((((((((.......))))))))..........))))))))).)))).. ( -24.80, z-score = -2.19, R) >droSec1.super_0 6721600 110 - 21120651 ------AAUCACAAGCAAUGGCAAUGGUGCAAUAA-UUUUCUGUGGGCGAGCGGAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAA ------...(((..((....))....)))((((.(-((((((((...(.(((......))).).....((((((((.......))))))))..........))))))))).)))).. ( -24.80, z-score = -2.19, R) >droYak2.chr3L 14684788 116 - 24197627 AAUCACAAUCACAAGCAACGGCAAUGGUGCAAUAAUUUUUUUGUGGGCGAGC-GAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAA .....((((((....((((.(((....))).......(((((((......))-)))))))))......((((((((.......))))))))..................)))))).. ( -26.40, z-score = -2.11, R) >droEre2.scaffold_4784 14566150 108 - 25762168 ------AAUCACAAGC-ACGGCAAUGGUGCAAUAA-UUUUUUGUGGGCGAGC-GAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAA ------(((((...((-((.......))))..(((-((((((((......))-)))))))))......((((((((.......))))))))..................)))))... ( -26.00, z-score = -2.80, R) >droWil1.scaffold_181136 730594 91 + 2313701 ------------------UCACAGUUGCACAA-------AUUGCAGAAUAGG-UAGAGAUGUAAAAAGUGUCAGCAAAAUAAAUGCUGACGAUAAAUAAAAAUGGAAAAUGAUUGAA ------------------...((((((((...-------..)))........-...............((((((((.......))))))))...................))))).. ( -15.20, z-score = -1.61, R) >consensus ______AAUCACAAGCAAUGGCAAUGGUGCAAUAA_UUUUUUGUGGGCGAGC_GAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAA .............................((((...((((((((...(((.(.....).)))......((((((((.......))))))))..........))))))))..)))).. (-15.40 = -15.65 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:17 2011