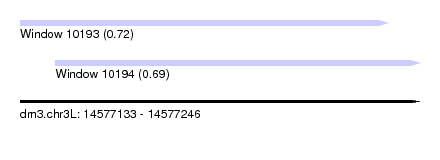
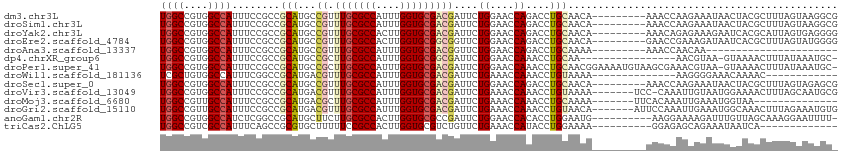
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,577,133 – 14,577,246 |
| Length | 113 |
| Max. P | 0.720054 |

| Location | 14,577,133 – 14,577,237 |
|---|---|
| Length | 104 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.59434 |
| G+C content | 0.51612 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14577133 104 + 24543557 UGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACA---------AAACCAAGAAAUAACUACGCUUUAGUAAGGCG ((((....))))....(((((((...((((.(((((....)))))))))..((((....))))..)))....---------.............((((.....))))..)))) ( -32.50, z-score = -1.39, R) >droSim1.chr3L 13902951 104 + 22553184 UGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACA---------AAACCAAGAAAUAACUACGCUUUAGUAAGGCG ((((....))))....(((((((...((((.(((((....)))))))))..((((....))))..)))....---------.............((((.....))))..)))) ( -32.50, z-score = -1.39, R) >droYak2.chr3L 14664874 104 + 24197627 UGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCACUUGGUGCGACGAUUCUGGAACCAGACCUGCAACA---------AAACAGAGAAAGAAUCACGCAUUAGUGAGGGG ((((....))))...((.(((((...((((.(((((....)))))))))..((((....))))..)))....---------..............((((......)))))))) ( -32.70, z-score = -0.53, R) >droEre2.scaffold_4784 14546946 104 + 25762168 UGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCACUUGGUGCGGCGGUUCUGGAACCAGACCUGCAACA---------GAACCGAAAGAUAAUCACGCUUUAGUAUGGGG ((((....))))......((.(((((.(((((((((....))))).((((.((((....)))).))))....---------))))(....)..............))))).)) ( -36.80, z-score = -0.63, R) >droAna3.scaffold_13337 18778742 82 - 23293914 UGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGGUUCUGGAACCAGACCUGCAAAA---------AAACCAACAA---------------------- ((((....))))........(((.((((((.(((((....)))))))))))((((....))))..)))....---------..........---------------------- ( -29.30, z-score = -1.43, R) >dp4.chrXR_group6 7275147 95 + 13314419 UGGCCGUGGCCAUUUCCGCCGCAUGCCGCUUGCGCCAUUUGGUGCGGCGAUUCUGGAACCAAACCUGCAA----------------AACGUAA-GUAAAACUUUAUAAAUGC- ((((((((((.......)))))....((((.(((((....))))))))).....))..))).....(((.----------------.....((-(.....)))......)))- ( -26.00, z-score = 0.31, R) >droPer1.super_41 631631 111 + 728894 UGGCCGUGGCCAUUUCCGCCGCAUGCCGCUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAAACCUGCAACGGAAAAUGUAAGCGAAACGUAA-GUAAAACUUUAUAAAUGC- ((((....))))((((((..(((...((.(((((((....)))))))))....((....))....)))..)))))).((((((....((....-)).....)))))).....- ( -30.10, z-score = -0.15, R) >droWil1.scaffold_181136 637756 87 - 2313701 UCGCUGUGGCCAUUUCGGCCGCAUGACGUUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGUAAAA--------------AAGGGGAAACAAAAC------------ (((.(((((((.....))))))))))((((.(((((....)))))))))..............(((......--------------.)))(....).....------------ ( -29.40, z-score = -2.43, R) >droSec1.super_0 6702121 104 + 21120651 UGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACA---------AAACCAAGAAAUAACUACGCUUUAGUAGAGCG (((..(((((.......))))).(((((((.(((((....)))))))))..((((....))))...)))...---------...)))...........(((((.....))))) ( -32.40, z-score = -1.73, R) >droVir3.scaffold_13049 16664084 105 + 25233164 UGGCCGUGGCCAUUUCCGCCGCAUGACGUUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGUAAAA-------UCC-CAAAUUGUAAUGGAAAACUUUAGCAAUGCG ((((....)))).......(((((..((((.(((((....))))))))).......................-------..(-((........)))............))))) ( -24.80, z-score = -0.05, R) >droMoj3.scaffold_6680 4714570 92 - 24764193 UGGCCGUUGCCAUUUCCGCCGCAUGACGCUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGCAAAA-------UUCACAAAUUGAAAUGGUAA-------------- ......((((((((((....(((.(.((.(((((((....)))))))))....((....))..).)))..((-------((....))))))))))))))-------------- ( -22.70, z-score = -0.34, R) >droGri2.scaffold_15110 8119635 106 - 24565398 UGGCCGUUGCCAUUUCCGCCGCAUGACGUUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGUAACA-------AUUCCAAAUUGAAAUGGCAAACUUUAGAAAUGUG ((((....)))).......(((((..((((.(((((....))))))))).(((((((......((.....((-------((.....))))....)).....)))))))))))) ( -24.60, z-score = -0.43, R) >anoGam1.chr2R 21283785 102 + 62725911 UGGCCGUGGCCAUCUCGGCCGCAUGCUUCUUGCGCCACUUGGUGCGCCGAUUCUGGAACCACACCUGGAAUG----------AAGGAAAAGAUUUGUUAGCAAAGGAAUUUU- ...((((((((.....)))))).(((((((((((((....))))).((.((((..(........)..)))).----------..))..))))......))))..))......- ( -33.10, z-score = -0.83, R) >triCas2.ChLG5 11190476 90 - 18847211 UGGCCGUCGCCAUUUCAGCCGCGUGCUUUUUCCGCCACUUGGUGCGUCUGUUCUGAAACCAUACCUGGAAAA----------GGAGAGCAGAAAUAAUCA------------- ((((....))))........((...(((((((((.....((((...((......)).))))....)))))))----------))...))...........------------- ( -23.40, z-score = -0.04, R) >consensus UGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAAACCUGCAACA_________AAACAAAGAAAAAACAAAACUUUAGUAAUGC_ ((((....))))........(((...((.(((((((....)))))))))....((....))....)))............................................. (-16.29 = -16.57 + 0.28)
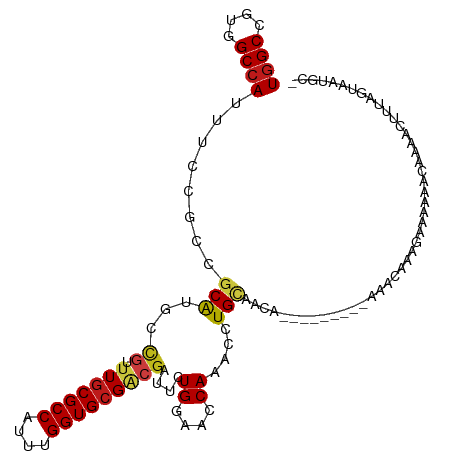

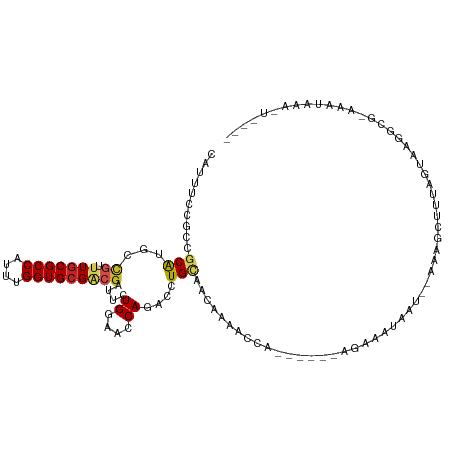
| Location | 14,577,143 – 14,577,246 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.38 |
| Shannon entropy | 0.63738 |
| G+C content | 0.46683 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -10.45 |
| Energy contribution | -10.79 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14577143 103 + 24543557 CAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACAAAACCA------AGAAAUAAC--UACGCUUUAGUAAGGCG-AAAUAAAAU---- .(((((.((((((...((((.(((((....)))))))))..((((....))))..)))..........------.......((--((.....))))..))))-)))).....---- ( -28.00, z-score = -2.41, R) >droSim1.chr3L 13902961 103 + 22553184 CAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACAAAACCA------AGAAAUAAC--UACGCUUUAGUAAGGCG-AAAUAAAAU---- .(((((.((((((...((((.(((((....)))))))))..((((....))))..)))..........------.......((--((.....))))..))))-)))).....---- ( -28.00, z-score = -2.41, R) >droYak2.chr3L 14664884 104 + 24197627 CAUUUCCGCCGCAUGCCGUUUGCGCCACUUGGUGCGACGAUUCUGGAACCAGACCUGCAACAAAACAG------AGAAAGAAU--CACGCAUUAGUGAGGGGCAAAUAAAAU---- .......(((.(.(((((((.(((((....)))))))))..((((....))))...))).........------........(--(((......))))).))).........---- ( -29.90, z-score = -1.59, R) >droEre2.scaffold_4784 14546956 104 + 25762168 CAUUUCCGCCGCAUGCCGUUUGCGCCACUUGGUGCGGCGGUUCUGGAACCAGACCUGCAACAGAACCG------AAAGAUAAU--CACGCUUUAGUAUGGGGCAAGUAAAAU---- .......(((.(((((.(((((((((....))))).((((.((((....)))).))))....))))(.------...).....--.........))))).))).........---- ( -35.80, z-score = -1.88, R) >dp4.chrXR_group6 7275157 94 + 13314419 CAUUUCCGCCGCAUGCCGCUUGCGCCAUUUGGUGCGGCGAUUCUGGAACCAAACCUGCAA----------------AACGUAAGUAAAACUUUAUAAAUGCCAGGGAAAU------ .(((((((((((.....))..(((((....))))))))....((((..............----------------.((....))...............))))))))))------ ( -27.31, z-score = -1.17, R) >droPer1.super_41 631641 110 + 728894 CAUUUCCGCCGCAUGCCGCUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAAACCUGCAACGGAAAAUGUAAGCGAAACGUAAGUAAAACUUUAUAAAUGCCAGGAAAAU------ ..(((((...((((..((((((((((....))))).....(((((...............))))).....)))))..((....))............))))..)))))..------ ( -28.06, z-score = -1.01, R) >droSec1.super_0 6702131 103 + 21120651 CAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACAAAACCA------AGAAAUAAC--UACGCUUUAGUAGAGCG-AAAUAAAAU---- .(((((.((.(((...((((.(((((....)))))))))..((((....))))..)))..........------........(--(((......)))).)))-)))).....---- ( -27.10, z-score = -2.45, R) >droVir3.scaffold_13049 16664094 95 + 25233164 CAUUUCCGCCGCAUGACGUUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGUAA-AAUCCCA------AAUUGUAAUGGAAAACUUUAGCAAUGCG-------------- .........(((((..((((.(((((....))))))))).....................-....(((------........)))............)))))-------------- ( -18.60, z-score = -0.22, R) >droGri2.scaffold_15110 8119645 96 - 24565398 CAUUUCCGCCGCAUGACGUUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGUAACAAUUCCA------AAUUGAAAUGGCAAACUUUAGAAAUGUG-------------- .........(((((..((((.(((((....))))))))).(((((((......((.....((((....------.))))....)).....))))))))))))-------------- ( -20.50, z-score = -0.87, R) >anoGam1.chr2R 21283795 96 + 62725911 CAUCUCGGCCGCAUGCUUCUUGCGCCACUUGGUGCGCCGAUUCUGGAACCACACCUGGAA------------------UGAAGGAAAAGAUUUGUUA--GCAAAGGAAUUUUCAAA ....(((((.(....).....(((((....))))))))))(((..(........)..)))------------------.....(((((..((((...--.))))....)))))... ( -23.90, z-score = 0.13, R) >triCas2.ChLG5 11190486 80 - 18847211 CAUUUCAGCCGCGUGCUUUUUCCGCCACUUGGUGCGUCUGUUCUGAAACCAUACCUGGAAAAGGAGAGC-----AGAAAUAAU--CA----------------------------- .(((((....((...(((((((((.....((((...((......)).))))....)))))))))...))-----.)))))...--..----------------------------- ( -19.70, z-score = -0.59, R) >consensus CAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACAAAACCA______AGAAAUAAU__AAAGCUUUAGUAAGGCG_AAAUAAA_U____ ..........(((...((.(((((((....)))))))))....((....))....))).......................................................... (-10.45 = -10.79 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:15 2011