| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,240,766 – 5,240,850 |
| Length | 84 |
| Max. P | 0.934624 |

| Location | 5,240,766 – 5,240,850 |
|---|---|
| Length | 84 |
| Sequences | 11 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Shannon entropy | 0.44357 |
| G+C content | 0.57995 |
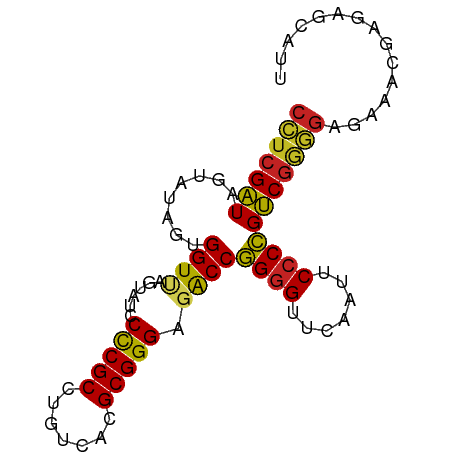
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -26.52 |
| Energy contribution | -25.90 |
| Covariance contribution | -0.62 |
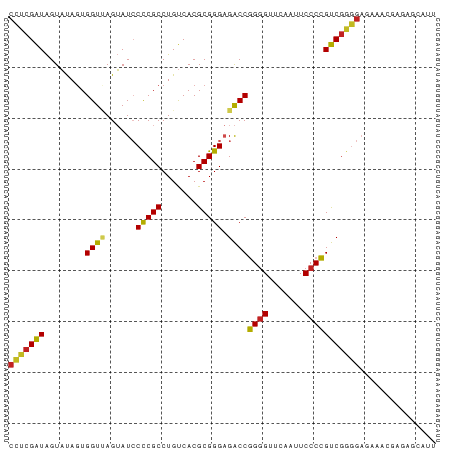
| Combinations/Pair | 1.35 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
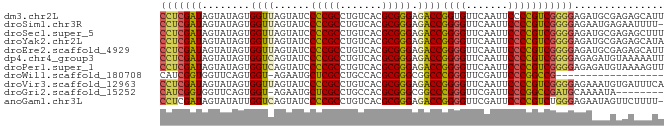
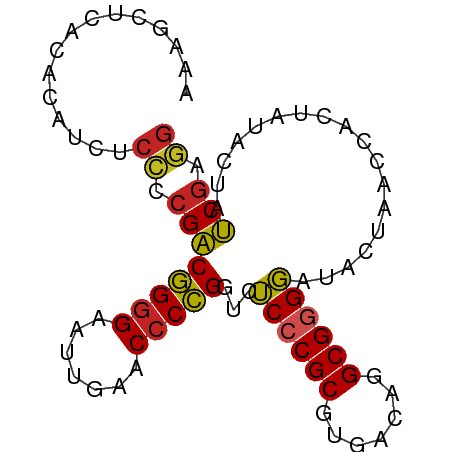
>dm3.chr2L 5240766 84 + 23011544 CCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGUGUUCAAUUCCCCGUCGGGGAGAUGCGAGAGCAUU (((((((......((((.........)))).((.(((.(((....))))))..)).......))))))).(((((....))))) ( -29.80, z-score = -1.42, R) >droSim1.chr3R 6940864 83 - 27517382 CCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAAUGAGAAUUUU- (((((((........((((......(((((.......))))).))))((((.......)))))))))))..............- ( -28.10, z-score = -0.81, R) >droSec1.super_5 3321267 84 + 5866729 CCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGCGAGAGCUUU (((((((........((((......(((((.......))))).))))((((.......)))))))))))....((....))... ( -31.30, z-score = -1.06, R) >droYak2.chr2L 8367253 84 - 22324452 CCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGCGAGAGCAUA (((((((........((((......(((((.......))))).))))((((.......)))))))))))..((((....)))). ( -33.90, z-score = -1.94, R) >droEre2.scaffold_4929 5319312 84 + 26641161 CCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGCGAGAGCAUU (((((((........((((......(((((.......))))).))))((((.......))))))))))).(((((....))))) ( -34.40, z-score = -2.17, R) >dp4.chr4_group3 7029977 84 + 11692001 CCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAUGUAAAAAUU (((((((........((((......(((((.......))))).))))((((.......)))))))))))............... ( -29.90, z-score = -1.52, R) >droPer1.super_1 8481897 84 + 10282868 CCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAUGUAAAAGUU (((((((........((((......(((((.......))))).))))((((.......)))))))))))............... ( -29.90, z-score = -1.47, R) >droWil1.scaffold_180708 11565966 65 - 12563649 CAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCG------------------ ........((((.(((((-((.........)))))))..))))((((.(((.......))))))).------------------ ( -26.70, z-score = -0.28, R) >droVir3.scaffold_12963 14507927 84 - 20206255 CCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAAAUGUGAUUUCA (((((((........((((......(((((.......))))).))))((((.......))))))))))).(((((...))))). ( -29.10, z-score = -0.99, R) >droGri2.scaffold_15252 5722934 75 - 17193109 CAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAAUA-------- ((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))).......-------- ( -31.00, z-score = -0.94, R) >anoGam1.chr3L 4285964 83 + 41284009 CCUCGAUAGUAUAUUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCGAUUCCCCGUCUGGGAGAAUAGUUCUUUU- (((.(((........((((......(((((.......))))).))))((((.......))))))).)))..............- ( -24.90, z-score = 0.14, R) >consensus CCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAAACGAGAGCAUU (((((((........((((......(((((.......))))).))))((((.......)))))))))))............... (-26.52 = -25.90 + -0.62)
| Location | 5,240,766 – 5,240,850 |
|---|---|
| Length | 84 |
| Sequences | 11 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Shannon entropy | 0.44357 |
| G+C content | 0.57995 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.06 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5240766 84 - 23011544 AAUGCUCUCGCAUCUCCCCGACGGGGAAUUGAACACCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGG ......((((((..(((((...)))))..)).......(((.(((((.......)))))))).................)))). ( -24.70, z-score = -0.48, R) >droSim1.chr3R 6940864 83 + 27517382 -AAAAUUCUCAUUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGG -...............((((((((((.......)))))(((.(((((.......))))))))................))).)) ( -24.90, z-score = -0.71, R) >droSec1.super_5 3321267 84 - 5866729 AAAGCUCUCGCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGG ...((....)).....((((((((((.......)))))(((.(((((.......))))))))................))).)) ( -26.00, z-score = -0.46, R) >droYak2.chr2L 8367253 84 + 22324452 UAUGCUCUCGCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGG .((((....))))...((((((((((.......)))))(((.(((((.......))))))))................))).)) ( -27.10, z-score = -0.80, R) >droEre2.scaffold_4929 5319312 84 - 26641161 AAUGCUCUCGCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGG .((((....))))...((((((((((.......)))))(((.(((((.......))))))))................))).)) ( -27.20, z-score = -0.86, R) >dp4.chr4_group3 7029977 84 - 11692001 AAUUUUUACAUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGG ................(((((.((((.......))))((((((((((.......)))))).....)))).........))).)) ( -25.70, z-score = -0.84, R) >droPer1.super_1 8481897 84 - 10282868 AACUUUUACAUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGG ................(((((.((((.......))))((((((((((.......)))))).....)))).........))).)) ( -25.70, z-score = -0.85, R) >droWil1.scaffold_180708 11565966 65 + 12563649 ------------------CGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUG ------------------((((((((.......)))))((((((.((...)).)))).)).....-............)))... ( -23.10, z-score = -0.33, R) >droVir3.scaffold_12963 14507927 84 + 20206255 UGAAAUCACAUUUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGG .(((((...)))))..((((((((((.......)))))(((.(((((.......))))))))................))).)) ( -25.20, z-score = -0.75, R) >droGri2.scaffold_15252 5722934 75 + 17193109 --------UAUUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUG --------.......(((((((((((.......)))))((((((.((...)).)))).)).....-............)))))) ( -27.80, z-score = -1.38, R) >anoGam1.chr3L 4285964 83 - 41284009 -AAAAGAACUAUUCUCCCAGACGGGGAAUCGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCAAUAUACUAUCGAGG -............(((......((((.......))))((((((((((.......)))))).....))))...........))). ( -24.50, z-score = -0.45, R) >consensus AAAGCUCACACAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGG ...............((.((((((((.......)))))...((((((.......))))))..................))).)) (-21.56 = -21.06 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:25 2011