| Sequence ID | dm3.chr2L |
|---|---|
| Location | 570,866 – 570,986 |
| Length | 120 |
| Max. P | 0.650999 |

| Location | 570,866 – 570,986 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Shannon entropy | 0.42092 |
| G+C content | 0.57222 |
| Mean single sequence MFE | -47.99 |
| Consensus MFE | -20.74 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650999 |
| Prediction | RNA |
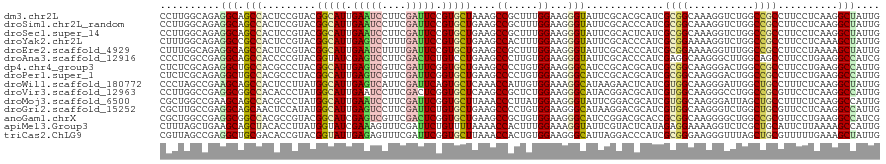
Download alignment: ClustalW | MAF
>dm3.chr2L 570866 120 + 23011544 CCUUGGCAGAGGCAGCCACUCCGUACGGCAUUGAAUCCUUCGAUUCCGUGCUAAAGCCGCUUUGGAAGGGUAUUCGCACGCAUCGCGGCAAAGGUCUGGCCGCCUUCCUCAAGGCUAUUG ((((((..(((((.((((..((((((((.(((((.....))))).))))))....(((((..((...(.(....).)...))..)))))...))..)))).)))))..))))))...... ( -50.40, z-score = -3.08, R) >droSim1.chr2L_random 29549 120 + 909653 CCUUGGCAGAGGCAGCCACUCCGUACGGCAUUGAAUCCUUCGAUUCCGUGCUGAAGCCGCUUUGGAAGGGUAUUCGCACCCAUCGCGGCAAAGGUCUGGCCGCCUUCCUCAAGGCUAUUG ((((((..(((((.((((..((...((((((.(((((....))))).))))))..(((((....((.((((......)))).)))))))...))..)))).)))))..))))))...... ( -55.40, z-score = -4.32, R) >droSec1.super_14 553559 120 + 2068291 CCUUGGCAGAGGCAGCCACUCCGUACGGCAUUGAAUCCUUCGAUUCCGUGCUGAAGCCGCUUUGGAAGGGUAUUCGCACUCAUCGCGGCAAAGGUCUGGCCGCCUUCCUCAAGGCUAUUG ((((((..(((((.((((..((...((((((.(((((....))))).))))))..(((((....((.((((......)))).)))))))...))..)))).)))))..))))))...... ( -52.40, z-score = -3.63, R) >droYak2.chr2L 559254 120 + 22324452 CUUUGGCAGAGGCCGCCACUCCGUACGGCAUUGAGUCCUUUGAUUCCGUGCUGAAGCCACUUUGGAAGGGUAUUCGCACCCAUCGCGGAAAAGGUCUGGCCGCCUUCCUCAAAGCUAUUG ((((((..(((((.((((..((...((((((.(((((....))))).)))))).......((((((.((((......)))).)).))))...))..)))).)))))..))))))...... ( -45.20, z-score = -2.23, R) >droEre2.scaffold_4929 626362 120 + 26641161 CUUUGGCAGAGGCAGCCACUCCGUACGGCAUUGAAUCUUUUGAUUCCGUGCUGAAGCCGCUUUGGAAGGGUAUUCGCACCCAUCGCGGAAAAGGUUUGGCCGCCUUCCUAAAAGCUAUUG .(((((..(((((.((((..((...((((((.(((((....))))).))))))...((((....((.((((......)))).))))))....))..)))).))))).)))))........ ( -46.50, z-score = -2.79, R) >droAna3.scaffold_12916 1572978 120 + 16180835 CCCUCGCCGAGGCAGCCACCCCGUACGGUAUCGAGUCCUUCGACUCUGUCCUGAAGCCCUUGUGGAAGGGUAUUCGCACCCAUCGAGGCAAGGGCUUGGCAGCCUUCCUGAAGGCCAUCG ((.(((..(((((.(((........(((.((.(((((....))))).)).)))((((((((((.((.((((......)))).))...))))))))))))).)))))..))).))...... ( -48.20, z-score = -1.84, R) >dp4.chr4_group3 10672548 120 - 11692001 CUCUCGCAGAGGCUGCCACGCCCUACGGCAUUGAGUCGUUCGAUUCGGUGCUGAAGCCCCUGUGGAAGGGCAUCCGCACGCAUCGCGGCAAGGGACUGGCCGCCUUCCUGAAGGCCAUUG (..(((..(((((.((((..((((.((((((((((((....))))))))))))..((((........))))..((((.......))))..))))..)))).)))))..)))..)...... ( -53.90, z-score = -1.88, R) >droPer1.super_1 7785992 120 - 10282868 CUCUCGCAGAGGCUGCCACGCCCUACGGCAUUGAGUCGUUCGAUUCGGUGCUGAAGCCCCUGUGGAAGGGCAUCCGCACGCAUCGCGGCAAGGGACUGGCCGCCUUCCUGAAGGCCAUUG (..(((..(((((.((((..((((.((((((((((((....))))))))))))..((((........))))..((((.......))))..))))..)))).)))))..)))..)...... ( -53.90, z-score = -1.88, R) >droWil1.scaffold_180772 6233134 120 - 8906247 CCCUAGCCGAAGCAGCCACUCCUUAUGGCAUUGAGUCAUUCGAUUCAGUGCUCAAACCAUUGUGGAAAGGCAUAAGAACUCAUCGUGGCAAGGGAUUGGCUGCCUUUCUCAAGGCUAUUG ...(((((((((((((((.(((((.((((((((((((....)))))))))).))..((((.((((..............)))).))))..))))).)))))))..)))....)))))... ( -44.84, z-score = -3.31, R) >droVir3.scaffold_12963 19023267 120 + 20206255 CCUUGGCCGAGGCGGCCACACCCUAUGGCAUUGAAUCCUUCGACUCGGUGCUCAAGCCGCUCUGGAAGGGCAUACGGACGCAUCGUGGCAAGGGCCUGGCCGCGUUCCUCAAGGCCAUUG ...(((((((((((((((..((((.(((((((((.((....)).))))))).)).(((((((.....))))..((((.....))))))).))))..)))))))....)))..)))))... ( -52.30, z-score = -1.72, R) >droMoj3.scaffold_6500 8187299 120 - 32352404 CGCUGGCCGAAGCAGCCACGCCCUAUGGCAUUGAAUCCUUCGAUUCGGUGCUUAAACCCUUAUGGAAGGGUAUUCGGACGCAUCGUGGCAAGGGAUUAGCUGCCUUUCUCAAGGCCAUUG ...(((((((((((((....((((..(((((((((((....)))))))))))...((((((....))))))........((......)).))))....)))))..)))....)))))... ( -44.20, z-score = -1.73, R) >droGri2.scaffold_15252 6923312 120 + 17193109 CGCUUGCCGAGGCAGCAACUCCAUAUGGCAUUGAGUCCUUCGAUUCUGUGCUGAAGCCCCUGUGGAAGGGCAUAAGGACGCAUCGUGGCAAGGGUCUGGCUGCGUUCCUCAAGGCCAUUG .(((.((....)))))........(((((.(((((((((((.((...(.((....)))...)).)))))))....((((((((((.(((....)))))).))))))).)))).))))).. ( -42.80, z-score = 0.00, R) >anoGam1.chrX 2990040 120 + 22145176 CGCUGGCCGAGGCGGCCACGCCGUACGGCAUCGAGUCGUUCGACUCGGUGCUGAAGCCGCUGUGGAAGGGCAUCCGGACGCACCGCGGCAAGGGGCUGGCCGCGUUCCUGAAGGCCAUCG ((.(((((((.(((((((.(((...((((((((((((....))))))))))))..(((((((((...((....))...))))..)))))....)))))))))).))......))))).)) ( -68.40, z-score = -3.32, R) >apiMel3.Group3 6794646 120 - 12341916 CUUUAGCUGAAGCAGCUACACCUUAUGGUAUCGAAAGUUUCGAUUCUGUUUUAAAACCACUUUGGAAAGGUAUUCGUACUCAUAGAGGAAAAGGUCUCGCUGCAUUCUUAAAAGCCAUUG .....(((((((((((...(((((((((.(((((.....))))).))))..........(((((....((((....))))..)))))...)))))...))))).))).....)))..... ( -25.30, z-score = -0.24, R) >triCas2.ChLG9 4367729 120 + 15222296 CGUUAGCCGAGGCUGCGACACCGUACGGUAUUGAGAGUUUCGAUUCGGUGCUUAAACCACUGUGGAAGGGCAUUAGGACCCAUCGCGGGAAGGGUUUAGCUGCGUUUUUGAAAGCUAUUG ...(((((((((((.(((.(((....))).)))..))))))).((((((......))).....(((((.(((.(..(((((.((....)).)))))..).))).)))))))).))))... ( -36.10, z-score = -0.20, R) >consensus CCUUGGCAGAGGCAGCCACUCCGUACGGCAUUGAGUCCUUCGAUUCGGUGCUGAAGCCGCUGUGGAAGGGUAUUCGCACGCAUCGCGGCAAGGGUCUGGCCGCCUUCCUCAAGGCCAUUG ..........(((.(((.........(((((.(((((....))))).)))))....((.....))...))).............((((...........))))..........))).... (-20.74 = -20.34 + -0.40)
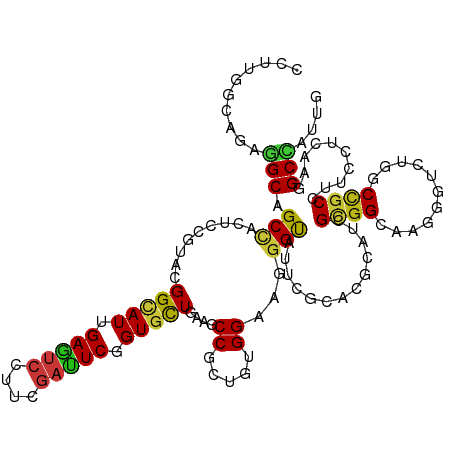
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:11 2011