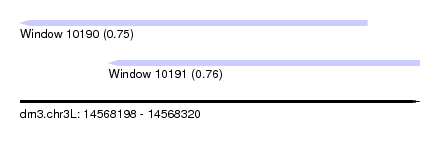
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,568,198 – 14,568,320 |
| Length | 122 |
| Max. P | 0.762615 |

| Location | 14,568,198 – 14,568,304 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.16 |
| Shannon entropy | 0.29914 |
| G+C content | 0.30890 |
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
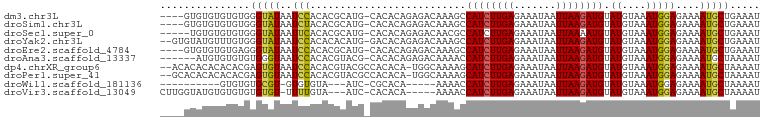
>dm3.chr3L 14568198 106 - 24543557 AAUCCACACGCAUG-CACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG------------- ..((((...((((.-....((...(((....((((((((.......))))))))..)))...))......)))).(((((.((((......)))))))))..)))).------------- ( -20.30, z-score = -1.35, R) >droSim1.chr3L 13894051 106 - 22553184 AAUCUACACGCAUG-CACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG------------- ..((((...((((.-....((...(((....((((((((.......))))))))..)))...))......)))).(((((.((((......)))))))))..)))).------------- ( -18.00, z-score = -0.50, R) >droSec1.super_0 6693229 106 - 21120651 AAUUCACACGCAUG-CACACAGAGACAACGCCAUCUUGAGAAAUAAUUAAAAUGUAUGUAAAUGGAGAAAAUGCUGAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG------------- ..(((....(((((-((....((((........)))).(....)........)))))))....((((((((((((............)))))....)))))))))).------------- ( -14.20, z-score = 0.76, R) >droYak2.chr3L 14655593 106 - 24197627 AAUCCACACACAUG-GACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG------------- ..((((........-....((...(((....((((((((.......))))))))..)))...))(((((((((((............)))))....)))))))))).------------- ( -19.80, z-score = -1.40, R) >droEre2.scaffold_4784 14537956 106 - 25762168 AAUCCACACGCAUG-CACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG------------- ..((((...((((.-....((...(((....((((((((.......))))))))..)))...))......)))).(((((.((((......)))))))))..)))).------------- ( -20.30, z-score = -1.35, R) >droAna3.scaffold_13337 18769089 105 + 23293914 -AUCCACACGUACG-CACACAGAGACAAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUCAGAG------------- -......(((((((-....)............(((((((.......)))))))))))))...(((((((((((((............)))))....))))))))...------------- ( -17.80, z-score = -1.71, R) >dp4.chrXR_group6 7265235 106 - 13314419 AAUCCACACGUACGCCACACA-UGGCAAAAGCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG------------- ..((((((..(((((((....-))))....(((((((((.......)))))))))..)))..))(((((((((((............)))))....)))))))))).------------- ( -27.10, z-score = -3.87, R) >droPer1.super_41 621683 106 - 728894 AAUCCACACGUACGCCACACA-UGGCAAAAGCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG------------- ..((((((..(((((((....-))))....(((((((((.......)))))))))..)))..))(((((((((((............)))))....)))))))))).------------- ( -27.10, z-score = -3.87, R) >droWil1.scaffold_181136 615190 91 + 2313701 -----------AAUCCGCACA-----AAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUAAGAG------------- -----------..........-----.......((((((((((((.......(.(((....))).)....(((((............)))))..)))))))))))).------------- ( -14.90, z-score = -2.13, R) >droVir3.scaffold_13049 16655284 95 - 25233164 -----------AAUCCACACA-----AAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUUGGCUACAG--------- -----------..........-----.....((((((((.......))))))))..((((..(..((((((((((............)))))....)))))..)..)))).--------- ( -17.60, z-score = -2.28, R) >droMoj3.scaffold_6680 4705190 104 + 24764193 -----------AAUCCACACA-----AAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUUGGCUACAGACCCAGCCC -----------..((((.(((-----.....((((((((.......))))))))..)))...))))(((((((((............)))))....))))...((((........)))). ( -18.10, z-score = -1.85, R) >droGri2.scaffold_15110 8110037 95 + 24565398 -----------AAUCCACACA-----AAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUUGGCUACCA--------- -----------..........-----.....((((((((.......))))))))...(((..(..((((((((((............)))))....)))))..)..)))..--------- ( -15.70, z-score = -1.50, R) >consensus AAUCCACACGCAUG_CACACA_AGACAAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAUUGUUAAAAGCAUAAUAUUUCUUUGGAG_____________ ...............................((((((((.......))))))))........(((((((((((((............)))))....))))))))................ (-13.59 = -13.55 + -0.04)


| Location | 14,568,225 – 14,568,320 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Shannon entropy | 0.39502 |
| G+C content | 0.35834 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -7.61 |
| Energy contribution | -7.47 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14568225 95 - 24543557 ----GUGUGUGUGUGGGUAUAAUCCACACGCAUG-CACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAU ----((((((((((((((...)))))))))))))-)...((...(((....((((((((.......))))))))..)))...))................ ( -29.30, z-score = -4.10, R) >droSim1.chr3L 13894078 95 - 22553184 ----GUGUGUGUGUGGGUAUAAUCUACACGCAUG-CACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAU ----((((((((((((((...)))))))))))))-)...((...(((....((((((((.......))))))))..)))...))................ ( -27.00, z-score = -3.46, R) >droSec1.super_0 6693256 94 - 21120651 -----UGUGUGUGUGGGUAUAAUUCACACGCAUG-CACACAGAGACAACGCCAUCUUGAGAAAUAAUUAAAAUGUAUGUAAAUGGAGAAAAUGCUGAAAU -----.((((((((((((...)))))))))))).-....(((........((((.(((....(((.........))).)))))))........))).... ( -17.89, z-score = -0.15, R) >droYak2.chr3L 14655620 97 - 24197627 --GUGUAUGUUUGUGGGUAUAAUCCACACACAUG-GACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAU --((.(((((.(((((((...))))))).)))))-.)).((...(((....((((((((.......))))))))..)))...))................ ( -24.80, z-score = -2.88, R) >droEre2.scaffold_4784 14537983 95 - 25762168 ----GUGUGUGUGAGGGUAUAAUCCACACGCAUG-CACACAGAGACAAAGCCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUGAAAU ----(((((((((.((((...)))).))))))))-)...((...(((....((((((((.......))))))))..)))...))................ ( -24.60, z-score = -2.85, R) >droAna3.scaffold_13337 18769116 93 + 23293914 ------AUGUGUGUGUGGGUAAUCCACACGUACG-CACACAGAGACAAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAU ------.((((((((((.((.......)).))))-))))))...(((....((((((((.......))))))))..)))..................... ( -23.30, z-score = -2.84, R) >dp4.chrXR_group6 7265262 97 - 13314419 --ACACACACACACGAGUGUAAUCCACACGUACGCCACACA-UGGCAAAAGCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAU --....((((......))))..((((....(((((((....-))))....(((((((((.......)))))))))..)))..)))).............. ( -23.20, z-score = -3.16, R) >droPer1.super_41 621710 97 - 728894 --GCACACACACACGAGUGUAAUCCACACGUACGCCACACA-UGGCAAAAGCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAU --(((.((((......))))..((((....(((((((....-))))....(((((((((.......)))))))))..)))..)))).....)))...... ( -25.60, z-score = -3.81, R) >droWil1.scaffold_181136 615217 80 + 2313701 ----------GUGUGUGCGU-GUGUGUA---AUC-CGCACA-----AAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAU ----------((.((((((.-.(.....---)..-))))))-----...))((((((((.......)))))))).......................... ( -16.60, z-score = -2.70, R) >droVir3.scaffold_13049 16655315 90 - 25233164 CUUGGUAUGUGUGUGUGUGU-UUUUGUA---AUC-CACACA-----AAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAU .(((((((((.((((((..(-(.....)---)..-))))))-----...))((((((((.......))))))))................)))))))... ( -20.30, z-score = -3.09, R) >consensus ____GUAUGUGUGUGGGUAUAAUCCACACGCAUG_CACACAGAGACAAAACCAUCUUGAGAAAUAAUUAAGAUGUAUGUAAAUGGAGAAAAUGCUAAAAU ...............(((((..((((...........(.....).......((((((((.......))))))))........))))....)))))..... ( -7.61 = -7.47 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:12 2011