| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,563,286 – 14,563,413 |
| Length | 127 |
| Max. P | 0.999653 |

| Location | 14,563,286 – 14,563,384 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 70.09 |
| Shannon entropy | 0.63765 |
| G+C content | 0.50639 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
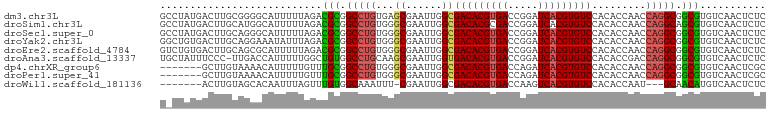
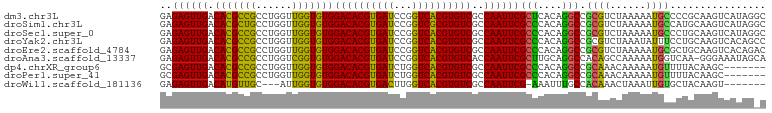
>dm3.chr3L 14563286 98 + 24543557 UGCCAGAGGAAAGAGUAUAUCCAACUGCCUAUGACUUGCGGGGCAUUUUUAGACGCGGCCUGUGAGCGAAUUGGCGACACGUGACCGGAUCACGUGUC .((((((((....(((.......))).))).((.((..((((.(............).))))..))))..)))))(((((((((.....))))))))) ( -33.20, z-score = -1.70, R) >droSim1.chr3L 13889231 98 + 22553184 UGCCAUAGGAAAGAGUAUAUCCAGCUGCCUAUGACUUGCAUGGCAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGCGACCGGAUCACGUGUC ..(((((((...(.((.........((((.(((.....))))))).......)).)..)))))))((......))((((((.((.....)).)))))) ( -28.79, z-score = 0.02, R) >droSec1.super_0 6688430 98 + 21120651 UGCCAAAGGAAAGAGUAUAUCCAGCUGCCUAUGACUUGCAGGGCAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUC .((((((((....(((.......))).))).((.((..((((.(............).))))..))))..)))))(((((((((.....))))))))) ( -34.40, z-score = -1.70, R) >droYak2.chr3L 14650275 98 + 24197627 UGACAGAAGCAAGGGUAUUCCCAGCUGGCUGUGACUUGCAGGAAAUAUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUC ..((((.(((..(((....))).)))..))))(.((..((((................))))..)))........(((((((((.....))))))))) ( -33.29, z-score = -1.29, R) >droEre2.scaffold_4784 14532908 98 + 25762168 UGCAAGAAGAAAGGGUAUACACAGCCGUCUGUGACUUGCAGCGCAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUC ((((((.(((...(((.......))).)))....))))))((((.......).))).(((.((......)).)))(((((((((.....))))))))) ( -31.00, z-score = -0.26, R) >droAna3.scaffold_13337 18764782 96 - 23293914 -UCUCGUUUAAAGAGUAUACCAAGUCUGCUAUUUCCCUU-GACCAUUUUUGGCUGUGGCCUGCAAGCGAAUUGGUGACACGUGACCGGAUCACGUGUC -.(((.......)))...(((((.((.(((.....((..-(.(((....))))...))......))))).)))))(((((((((.....))))))))) ( -26.80, z-score = -1.30, R) >dp4.chrXR_group6 6053976 91 - 13314419 -------UGCCGAAUCAAAGAAAAAAGAGUAGAGCUUGUAAAACAUUUUUGUUUGCGGCCUGUGGGCGAAUUGGCGACACGUGACCAGAUCACGUGUC -------.(((((.((....................((((((.((....))))))))(((....))))).)))))(((((((((.....))))))))) ( -29.80, z-score = -2.80, R) >droPer1.super_41 112038 91 - 728894 -------UGCCGAAUCAAGGAAAAAAGAGUAGAGCUUGUAAAACAUUUUUGUUUGCGGCCUGUGGGCGAAUUGGCGACACGUGACCAGAUCACGUGUC -------.(((((.((....................((((((.((....))))))))(((....))))).)))))(((((((((.....))))))))) ( -29.80, z-score = -2.53, R) >droWil1.scaffold_181136 1712443 81 + 2313701 ------------UACCAGGGCUAAA----UAAAACUUGUAGCACAAUUUAGUUUGUGGCAAAUUU-CGAAUUGGCGACACGUGACCAAGUCACGUGUC ------------.......(((((.----..(((.((((..(((((......))))))))).)))-....)))))((((((((((...)))))))))) ( -25.10, z-score = -3.06, R) >consensus UGCCAGAGGAAAGAGUAUACCCAGCUGCCUAUGACUUGCAGGACAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUC .........................................................(((.((......)).)))(((((((((.....))))))))) (-15.88 = -16.02 + 0.14)

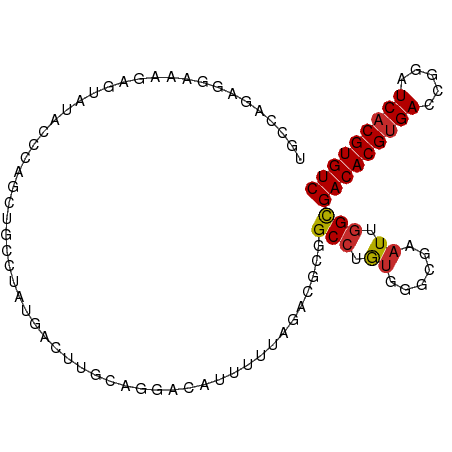

| Location | 14,563,286 – 14,563,384 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.09 |
| Shannon entropy | 0.63765 |
| G+C content | 0.50639 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.00 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14563286 98 - 24543557 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCUCACAGGCCGCGUCUAAAAAUGCCCCGCAAGUCAUAGGCAGUUGGAUAUACUCUUUCCUCUGGCA ((((((((((...))))))))))((((....(((....)))...((((((...((((.(....).....)))).)))))).............)))). ( -33.50, z-score = -2.88, R) >droSim1.chr3L 13889231 98 - 22553184 GACACGUGAUCCGGUCGCGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCAUGCAAGUCAUAGGCAGCUGGAUAUACUCUUUCCUAUGGCA ((((((((((...))))))))))((((....(((....)))...(((((....(((((((.....))).))))..))))).............)))). ( -33.80, z-score = -2.17, R) >droSec1.super_0 6688430 98 - 21120651 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCCUGCAAGUCAUAGGCAGCUGGAUAUACUCUUUCCUUUGGCA ((((((((((...))))))))))(((((...(((....))).(((((((....(((...))).....))))).)).(((.........))).))))). ( -33.10, z-score = -2.42, R) >droYak2.chr3L 14650275 98 - 24197627 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAUAUUUCCUGCAAGUCACAGCCAGCUGGGAAUACCCUUGCUUCUGUCA ((((((((((...))))))))))............(((((..(((......(((((((.((..((....))..)).)))))))....))).))))).. ( -28.60, z-score = -1.57, R) >droEre2.scaffold_4784 14532908 98 - 25762168 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCGCUGCAAGUCACAGACGGCUGUGUAUACCCUUUCUUCUUGCA ((((((((((...))))))))))((......(((....))).((((......)))))).(((((.(((((....))))).............))))). ( -32.44, z-score = -2.19, R) >droAna3.scaffold_13337 18764782 96 + 23293914 GACACGUGAUCCGGUCACGUGUCACCAAUUCGCUUGCAGGCCACAGCCAAAAAUGGUC-AAGGGAAAUAGCAGACUUGGUAUACUCUUUAAACGAGA- ((((((((((...))))))))))(((((.(((((......((...((((....)))).-...))....))).)).)))))...(((.......))).- ( -31.50, z-score = -3.01, R) >dp4.chrXR_group6 6053976 91 + 13314419 GACACGUGAUCUGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCAAACAAAAAUGUUUUACAAGCUCUACUCUUUUUUCUUUGAUUCGGCA------- ((((((((((...))))))))))(((.....(((....)))..((((.(((((.((............))...))))).))))....))).------- ( -25.60, z-score = -3.00, R) >droPer1.super_41 112038 91 + 728894 GACACGUGAUCUGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCAAACAAAAAUGUUUUACAAGCUCUACUCUUUUUUCCUUGAUUCGGCA------- ((((((((((...))))))))))(((...(((((....))).(((((((....))))).....)).................))...))).------- ( -25.50, z-score = -2.96, R) >droWil1.scaffold_181136 1712443 81 - 2313701 GACACGUGACUUGGUCACGUGUCGCCAAUUCG-AAAUUUGCCACAAACUAAAUUGUGCUACAAGUUUUA----UUUAGCCCUGGUA------------ ((((((((((...))))))))))((((....(-(((((((.(((((......)))))...)))))))).----........)))).------------ ( -25.52, z-score = -3.64, R) >consensus GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCCUGCAAGUCAUAGGCAGCUGGAUAUACUCUUUCAUCUGGCA ((((((((((...))))))))))........(((....))).((((......)))).......................................... (-17.53 = -17.00 + -0.53)


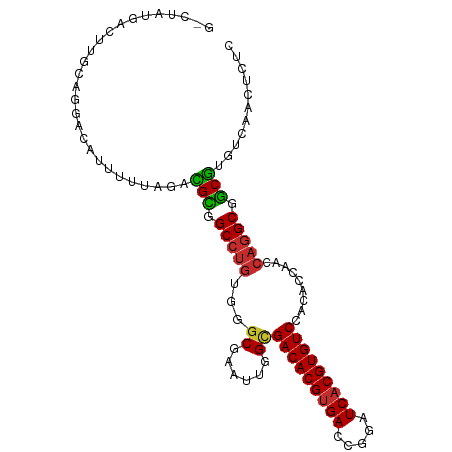
| Location | 14,563,312 – 14,563,413 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Shannon entropy | 0.37106 |
| G+C content | 0.56237 |
| Mean single sequence MFE | -35.51 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.71 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14563312 101 + 24543557 GCCUAUGACUUGCGGGGCAUUUUUAGACGCGGCCUGUGAGCGAAUUGGCGACACGUGACCGGAUCACGUGUCCACACCAACCAGGCGGCGUGUCAACUCUC ((((..........))))........((((.(((((.(.((......))(((((((((.....)))))))))........)))))).)))).......... ( -37.10, z-score = -1.20, R) >droSim1.chr3L 13889257 101 + 22553184 GCCUAUGACUUGCAUGGCAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGCGACCGGAUCACGUGUCCACACCAACCAGGCAGCGUGUCAACUCUC (((.(((.....)))))).......((((((((((((((((......))((((((.((.....)).))))))....)))..)))))..))))))....... ( -36.80, z-score = -1.62, R) >droSec1.super_0 6688456 101 + 21120651 GCCUAUGACUUGCAGGGCAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUCCACACCAACCAGGCGGCGUGUCAACUCUC ((((.((.....))))))........((((.((((((((((......))(((((((((.....)))))))))....)))..))))).)))).......... ( -40.20, z-score = -2.05, R) >droYak2.chr3L 14650301 101 + 24197627 GGCUGUGACUUGCAGGAAAUAUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUCCACACCAACCAGGCGGCGUGUCAACUCUC ..((((.....))))...........((((.((((((((((......))(((((((((.....)))))))))....)))..))))).)))).......... ( -38.40, z-score = -1.54, R) >droEre2.scaffold_4784 14532934 101 + 25762168 GUCUGUGACUUGCAGCGCAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUCCACACCAACCAGGCGGCGUGUCAACUCUC .....((((.(((...))).......((((.((((((((((......))(((((((((.....)))))))))....)))..))))).))))))))...... ( -39.00, z-score = -1.31, R) >droAna3.scaffold_13337 18764807 100 - 23293914 UGCUAUUUCCC-UUGACCAUUUUUGGCUGUGGCCUGCAAGCGAAUUGGUGACACGUGACCGGAUCACGUGUCCACACCGACCAGGCGGCGUGUCAACUCUC .((((.(((.(-(((.((((........))))....)))).))).))))(((((((((.....)))))))))(((.(((......))).)))......... ( -33.00, z-score = -0.99, R) >dp4.chrXR_group6 6054002 94 - 13314419 -------GCUUGUAAAACAUUUUUGUUUGCGGCCUGUGGGCGAAUUGGCGACACGUGACCAGAUCACGUGUCCACACCAACCAGGCGGCGUGUCAACUCGC -------((..((.(((((....)))))((.((((((((((......))(((((((((.....)))))))))....)))..))))).))......))..)) ( -34.70, z-score = -1.59, R) >droPer1.super_41 112064 94 - 728894 -------GCUUGUAAAACAUUUUUGUUUGCGGCCUGUGGGCGAAUUGGCGACACGUGACCAGAUCACGUGUCCACACCAACCAGGCGGCGUGUCAACUCGC -------((..((.(((((....)))))((.((((((((((......))(((((((((.....)))))))))....)))..))))).))......))..)) ( -34.70, z-score = -1.59, R) >droWil1.scaffold_181136 1712460 90 + 2313701 -------ACUUGUAGCACAAUUUAGUUUGUGGCAAAUUU-CGAAUUGGCGACACGUGACCAAGUCACGUGUCCACACCAAU---GCAACAUGUCAACUCUC -------..(((((((.((((((((((((...)))))).-.))))))))((((((((((...))))))))))........)---))))............. ( -25.70, z-score = -2.05, R) >consensus G_CUAUGACUUGCAGGACAUUUUUAGACGCGGCCUGUGGGCGAAUUGGCGACACGUGACCGGAUCACGUGUCCACACCAACCAGGCGGCGUGUCAACUCUC ...........................(((.(((((...((......))(((((((((.....))))))))).........))))).)))........... (-26.86 = -26.71 + -0.15)
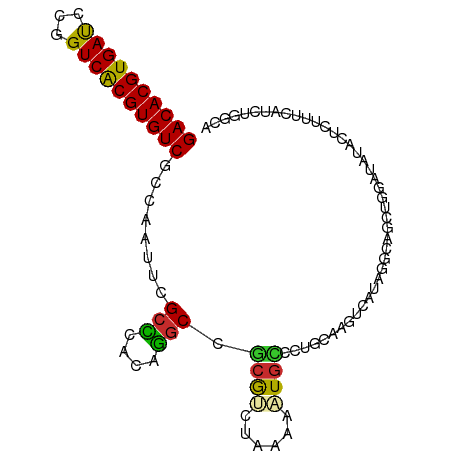
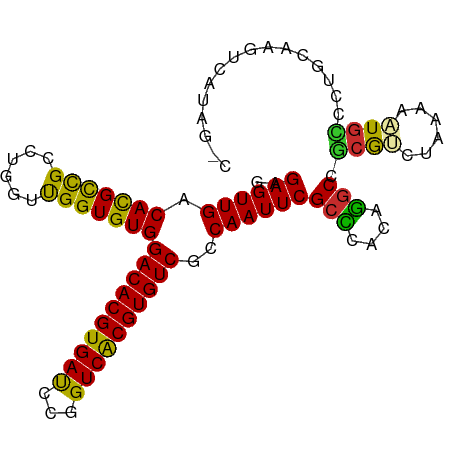
| Location | 14,563,312 – 14,563,413 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Shannon entropy | 0.37106 |
| G+C content | 0.56237 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -32.24 |
| Energy contribution | -31.24 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14563312 101 - 24543557 GAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCUCACAGGCCGCGUCUAAAAAUGCCCCGCAAGUCAUAGGC ......((((((((.(((((((((((...((((((((((...)))))))))))))))).......))))).)))).......(((...))).))))..... ( -37.71, z-score = -1.38, R) >droSim1.chr3L 13889257 101 - 22553184 GAGAGUUGACACGCUGCCUGGUUGGUGUGGACACGUGAUCCGGUCGCGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCAUGCAAGUCAUAGGC ......((((((((.(((((...((((.(((((((((((...)))))))))).).....))))..))))).)))).......(((...))).))))..... ( -38.10, z-score = -1.29, R) >droSec1.super_0 6688456 101 - 21120651 GAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCCUGCAAGUCAUAGGC ......((((((((.(((((...((((.(((((((((((...)))))))))).).....))))..))))).)))).......(((...))).))))..... ( -38.50, z-score = -1.63, R) >droYak2.chr3L 14650301 101 - 24197627 GAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAUAUUUCCUGCAAGUCACAGCC ..((.(((..((((.(((((...((((.(((((((((((...)))))))))).).....))))..))))).))))..............))).))...... ( -35.99, z-score = -1.45, R) >droEre2.scaffold_4784 14532934 101 - 25762168 GAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCGCUGCAAGUCACAGAC ....((.(((((((.(((((...((((.(((((((((((...)))))))))).).....))))..))))).)))).......(((...))).))))).... ( -38.50, z-score = -1.35, R) >droAna3.scaffold_13337 18764807 100 + 23293914 GAGAGUUGACACGCCGCCUGGUCGGUGUGGACACGUGAUCCGGUCACGUGUCACCAAUUCGCUUGCAGGCCACAGCCAAAAAUGGUCAA-GGGAAAUAGCA ..((((((.(((((((......)))))))((((((((((...))))))))))..))))))(((......((...((((....))))...-.))....))). ( -40.50, z-score = -2.61, R) >dp4.chrXR_group6 6054002 94 + 13314419 GCGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCUGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCAAACAAAAAUGUUUUACAAGC------- ((((((((.(((((((......)))))))((((((((((...))))))))))..))))))(((....))).))(((((....))))).......------- ( -37.00, z-score = -2.33, R) >droPer1.super_41 112064 94 + 728894 GCGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCUGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCAAACAAAAAUGUUUUACAAGC------- ((((((((.(((((((......)))))))((((((((((...))))))))))..))))))(((....))).))(((((....))))).......------- ( -37.00, z-score = -2.33, R) >droWil1.scaffold_181136 1712460 90 - 2313701 GAGAGUUGACAUGUUGC---AUUGGUGUGGACACGUGACUUGGUCACGUGUCGCCAAUUCG-AAAUUUGCCACAAACUAAAUUGUGCUACAAGU------- ..((((((.((..((..---...))..))((((((((((...))))))))))..)))))).-..(((((.(((((......)))))...)))))------- ( -28.20, z-score = -1.61, R) >consensus GAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCCUGCAAGUCAUAG_C ..((((((.(((((((......)))))))((((((((((...))))))))))..))))))(((....))).((((......))))................ (-32.24 = -31.24 + -0.99)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:11 2011