| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,561,778 – 14,561,829 |
| Length | 51 |
| Max. P | 0.601527 |

| Location | 14,561,778 – 14,561,829 |
|---|---|
| Length | 51 |
| Sequences | 11 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 88.88 |
| Shannon entropy | 0.21393 |
| G+C content | 0.43485 |
| Mean single sequence MFE | -13.80 |
| Consensus MFE | -8.63 |
| Energy contribution | -8.73 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14561778 51 + 24543557 UUUAUGC----UGCCGCAACCUGAACGAGGCAUAAAACAA-----AUGUCAUACUCAGGA ....(((----....))).(((((....(((((.......-----)))))....))))). ( -12.20, z-score = -2.14, R) >droSim1.chr3L 13887711 51 + 22553184 UUUAUGC----UGCCGCAACCUGAACGAGGCAUAAAACAA-----AUGUCAUACUCAGGA ....(((----....))).(((((....(((((.......-----)))))....))))). ( -12.20, z-score = -2.14, R) >droSec1.super_0 6686902 51 + 21120651 UUUAUGC----UGCCGCAACCUGAAGGAGGCAUAAAACAA-----AUGUCAUACUCAGGA ....(((----....))).(((((..(.(((((.......-----))))).)..))))). ( -12.50, z-score = -1.69, R) >droYak2.chr3L 14648686 51 + 24197627 UUUAUGC----UGCCGCAACCUGAACGAGGCAUAAAACAA-----AUGUCAUACUCAGGA ....(((----....))).(((((....(((((.......-----)))))....))))). ( -12.20, z-score = -2.14, R) >droEre2.scaffold_4784 14531433 51 + 25762168 UUUAUGC----UGCCGCAACCUGAACGAGGCAUAAAACAA-----AUGUCAUACUCAGGA ....(((----....))).(((((....(((((.......-----)))))....))))). ( -12.20, z-score = -2.14, R) >droAna3.scaffold_13337 18763436 51 - 23293914 UUUAUGC----UGCCGCAACCUGAUCGGGGCAUAAAACAA-----AUGUCAUACUCAGGG ....(((----....))).(((((....(((((.......-----)))))....))))). ( -13.50, z-score = -1.94, R) >dp4.Unknown_singleton_3270 1476 51 + 1721 UUUAUGC----UGCCGCAACCUGAGGUUGGCAUAAAACAA-----AUGUCAUACUCAGGA ....(((----....))).((((((..((((((.......-----))))))..)))))). ( -18.60, z-score = -3.80, R) >droPer1.super_41 615721 51 + 728894 UUUAUGC----UGCCGCAACCUGAGGUUGGCAUAAAACAA-----AUGUCAUACUCAGGA ....(((----....))).((((((..((((((.......-----))))))..)))))). ( -18.60, z-score = -3.80, R) >droWil1.scaffold_181136 587898 60 - 2313701 UUUAUGCCGCAGGCUGCAACCUGAGGUAGGCAUAAAACAACACAAAUGUCAUACUUAGGG ((((((((.((((......)))).....))))))))........................ ( -14.70, z-score = -1.28, R) >droVir3.scaffold_13049 8584840 51 - 25233164 UUUAUGC----GACCGCAGCCUGAAGUAGGCAUAAAACAA-----AUGUCAUACUCAGAA ....(((----....)))..((((.((((((((.......-----))))).))))))).. ( -12.40, z-score = -2.45, R) >droGri2.scaffold_15110 16462829 51 + 24565398 UUUAUGC----GGCCACAGCCUGAAGUUGGCAUAAAACAA-----AUGUCAUACUCAGGA .......----........(((((.((((((((.......-----)))))).))))))). ( -12.70, z-score = -1.48, R) >consensus UUUAUGC____UGCCGCAACCUGAAGGAGGCAUAAAACAA_____AUGUCAUACUCAGGA ....(((........))).(((((....(((((............)))))....))))). ( -8.63 = -8.73 + 0.10)

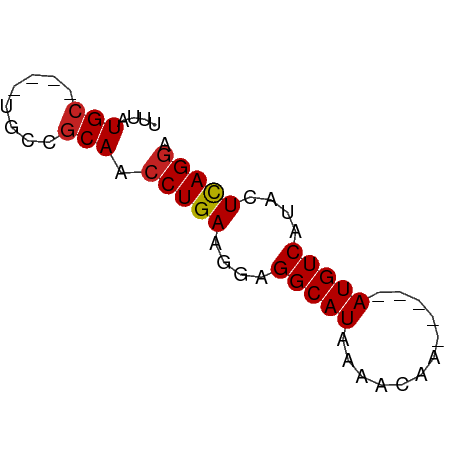

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:08 2011