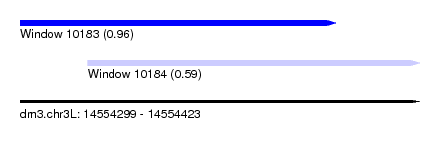
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,554,299 – 14,554,423 |
| Length | 124 |
| Max. P | 0.957700 |

| Location | 14,554,299 – 14,554,397 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.63 |
| Shannon entropy | 0.48917 |
| G+C content | 0.43149 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14554299 98 + 24543557 CCCCUUUUCUCCGCUCGC----------CACUUGU-ACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCC----- .((((((..((.((..((----------.......-.......))..)).))..)).))))..((((((((......))))))))........................----- ( -17.84, z-score = -1.14, R) >droSim1.chr3L 13880006 98 + 22553184 CCCCUUUUCUCCGCUCGC----------CACUUGU-ACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCC----- .((((((..((.((..((----------.......-.......))..)).))..)).))))..((((((((......))))))))........................----- ( -17.84, z-score = -1.14, R) >droSec1.super_0 6679617 98 + 21120651 CCCCUUUUCUCCGCUCGC----------CACUUGU-ACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCC----- .((((((..((.((..((----------.......-.......))..)).))..)).))))..((((((((......))))))))........................----- ( -17.84, z-score = -1.14, R) >droYak2.chr3L 14640573 98 + 24197627 CCCCUUUUCACCGCUCGC----------CACUUGU-GCAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCC----- ............((.((.----------....)).-))...((((..(((....)))..))))((((((((......))))))))........................----- ( -19.80, z-score = -1.75, R) >droEre2.scaffold_4784 14523954 103 + 25762168 CCCCUUUUCUCCGCUCGC----------CACUUGG-GCAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUUUCC ..................----------....(((-((...((((..(((....)))..))))((((((((......)))))))).......)))))................. ( -25.10, z-score = -2.90, R) >droAna3.scaffold_13337 18756910 98 - 23293914 CCCCUUUUUUGUGCUCGC----------CACUUGU-ACAAAUUGCUGGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUUGUCGACUUUUCUUUUCC----- .......(((((((....----------.....))-)))))(((((.(((....))).)))))((((((((......))))))))........................----- ( -23.50, z-score = -2.09, R) >droPer1.super_41 603655 107 + 728894 CCCCUUUUUGCCUGCCUUGCCACUCACUCAGUCGU-ACAAAU-GCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGCCACUUUUUCCGUUGC----- .......((((((((..((.....)).((((.((.-.(....-)..))))))...))))))))((((((((......)))))))).......((.((.......)).))----- ( -26.40, z-score = -2.46, R) >dp4.chrXR_group6 7250719 107 + 13314419 CCCCUUUUUGCCUGCCUUGCCACUCACUCAGUCGU-ACAAAU-GCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGCCACUUUUUCCGUUGC----- .......((((((((..((.....)).((((.((.-.(....-)..))))))...))))))))((((((((......)))))))).......((.((.......)).))----- ( -26.40, z-score = -2.46, R) >droWil1.scaffold_181136 508739 99 - 2313701 CCCUCUAAAGUCAGUAGU----------CACUUGGGACAAAUUGCUCGCUGAUUAGCAAGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUUGCAAACUUUCUCUUUCU----- ......(((((..(((((----------(......)))...(((((.(((....))).)))))((((((((......))))))))......)))..)))))........----- ( -23.80, z-score = -2.49, R) >droVir3.scaffold_13049 16637893 100 + 25233164 CCCUUUUGGGGCAGCCUUG----UCAGCCAGUUGU-ACAAAUUGCUCGCUGAUUAGCAGGCAAGCGCAUGAAAUUAAUCAUAUGCAAAUACGAGUAUGCUACCAC--------- (((....)))(((((((.(----(((((.(((.((-....)).))).))))))....))))..(((.((((......)))).)))...........)))......--------- ( -28.70, z-score = -1.16, R) >droMoj3.scaffold_6680 4685311 85 - 24764193 CCCUUUGUCGGCAGCCUUG----UCAGGCAGUUGU-ACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUA------------------------ ...(((((((((.((((..----..)))).)))).-)))))((((..(((....)))..))))((((((((......)))))))).....------------------------ ( -29.60, z-score = -3.28, R) >droGri2.scaffold_15110 8093102 92 - 24565398 CCCCUU----UCGGUUGG----------CAGUUGU-ACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUA--GCCAAGUAUCCUUCCAU----- ......----(((((.((----------(((((..-...))))))).)))))......(((..((((((((......)))))))).....--)))..............----- ( -26.20, z-score = -2.69, R) >consensus CCCCUUUUCGCCGCUCGC__________CACUUGU_ACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCAACUUUUCUUUUCC_____ .........................................(((((........)))))....((((((((......))))))))............................. (-12.29 = -12.47 + 0.17)
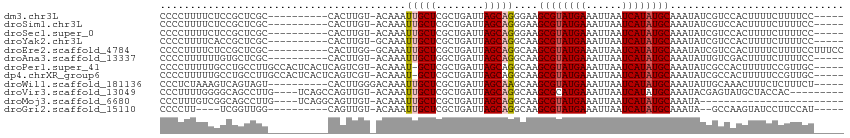
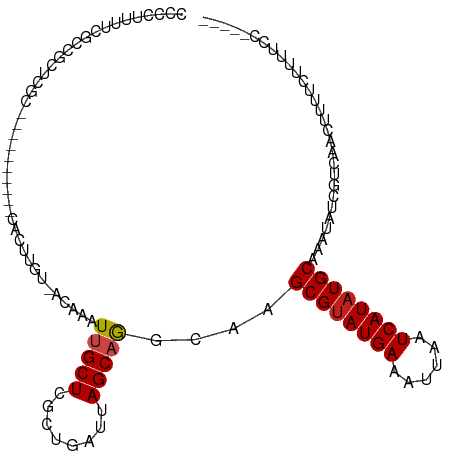
| Location | 14,554,320 – 14,554,423 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.62 |
| Shannon entropy | 0.50563 |
| G+C content | 0.40086 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -12.22 |
| Energy contribution | -12.41 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14554320 103 + 24543557 --------UUGUACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUUGACGC-----GAAUUUGUUGCUGUCGUGG --------..((((((((((((((((....)))((((((((((((((......)))))))).......(....).......)))))))).))-----.)))))).)))........ ( -24.00, z-score = -0.86, R) >droSim1.chr3L 13880027 103 + 22553184 --------UUGUACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUCGACGC-----GAAUUUGUUGCUGUCGUGG --------.((.((.......((.(((.....))).)).((((((((......)))))))).......)).))..........((.((((((-----((.....)))).)))).)) ( -25.20, z-score = -1.19, R) >droSec1.super_0 6679638 103 + 21120651 --------UUGUACAAAUUGCUCGCUGAUUAGCAGGGAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUCGACGC-----GAAUUUGUUGCUGUCGUGG --------.((.((.......((.(((.....))).)).((((((((......)))))))).......)).))..........((.((((((-----((.....)))).)))).)) ( -25.20, z-score = -1.19, R) >droYak2.chr3L 14640594 103 + 24197627 --------UUGUGCAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUUGACGC-----GAAUUUGUUGCUGUCGUGG --------.((..(...((((..(((....)))..))))((((((((......)))))))).......)..))..........((.((((((-----((.....)))).)))).)) ( -27.40, z-score = -1.63, R) >droEre2.scaffold_4784 14523975 108 + 25762168 --------UUGGGCAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUUUCCUUGAAGCGAAUUUGUUGCUGUCGUGG --------.(((((...((((..(((....)))..))))((((((((......)))))))).......)))))...............((.((((((((.....))))).))).)) ( -31.20, z-score = -2.51, R) >droAna3.scaffold_13337 18756931 105 - 23293914 --------UUGUACAAAUUGCUGGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUUGUCGACUUUUCUUUUCCUUGACGCC---CAAUGUUGUUGCUGUCGUGG --------(((.((((.(((((.(((....))).)))))((((((((......)))))))).....)))))))..........((.((((((.---((....))..)).)))).)) ( -25.40, z-score = -0.91, R) >droPer1.super_41 603678 104 + 728894 UCACUCAGUCGUACAAAU-GCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGCCACUUUUUCCGUUGCUC-----------AAGUUGUUGAUGUCCUUG .......((((.((((.(-((..(((....)))..))).((((((((......)))))))).......((.((.......)).))..-----------...))))))))....... ( -22.60, z-score = -0.75, R) >dp4.chrXR_group6 7250742 104 + 13314419 UCACUCAGUCGUACAAAU-GCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGCCACUUUUUCCGUUGCUC-----------AAGUUGUUGAUGUCCUUG .......((((.((((.(-((..(((....)))..))).((((((((......)))))))).......((.((.......)).))..-----------...))))))))....... ( -22.60, z-score = -0.75, R) >droWil1.scaffold_181136 508760 104 - 2313701 -------UUGGGACAAAUUGCUCGCUGAUUAGCAAGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUUG-CAAACUUUCUCUUUCUUUUUUUAU----AGCUUCUUUCUGUGUCGC -------....((((..(((((.(((....))).)))))((((((((......))))))))........-........................----............)))).. ( -22.30, z-score = -1.15, R) >droVir3.scaffold_13049 16637920 80 + 25233164 --------UUGUACAAAUUGCUCGCUGAUUAGCAGGCAAGCGCAUGAAAUUAAUCAUAUGCAAAUA----CGAGUAUGCUACCACUUUGCGC------------------------ --------..((((...((((..(((....)))..))))(((.((((......)))).))).....----...))))((.........))..------------------------ ( -17.00, z-score = 0.33, R) >droGri2.scaffold_15110 8093119 81 - 24565398 --------UUGUACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAGC--CAAGUAUCCUUCCAUUUGGCG------------------------- --------.........((((..(((....)))..))))((((((((......)))))))).....((--(((((........))))))).------------------------- ( -26.40, z-score = -4.04, R) >consensus ________UUGUACAAAUUGCUCGCUGAUUAGCAGGCAAGCGUAUGAAAUUAAUCAUAUGCAAAUAUCGUCCACUUUUCUUUUCCUUGACGC______AAUUUGUUGCUGUCGUGG .................(((((........)))))....((((((((......))))))))....................................................... (-12.22 = -12.41 + 0.19)

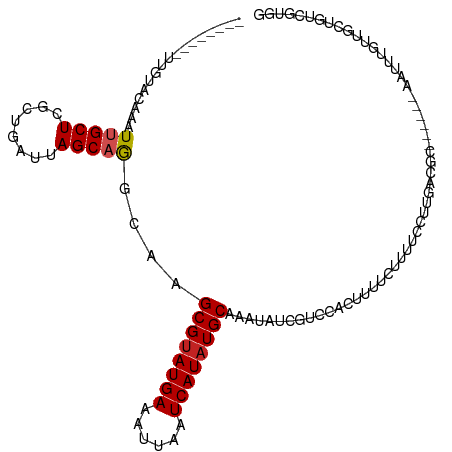
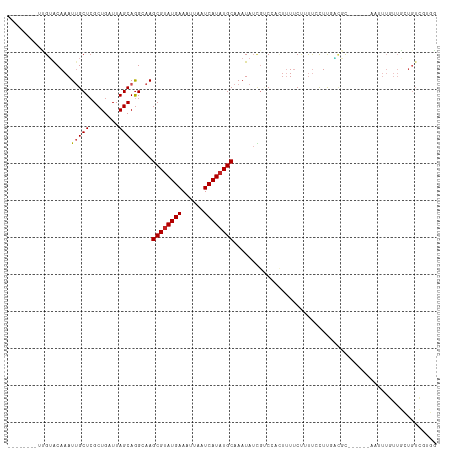
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:07 2011