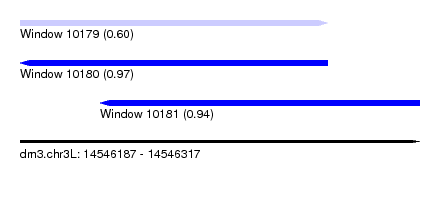
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,546,187 – 14,546,317 |
| Length | 130 |
| Max. P | 0.966627 |

| Location | 14,546,187 – 14,546,287 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.41 |
| Shannon entropy | 0.63615 |
| G+C content | 0.43275 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -8.74 |
| Energy contribution | -9.39 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
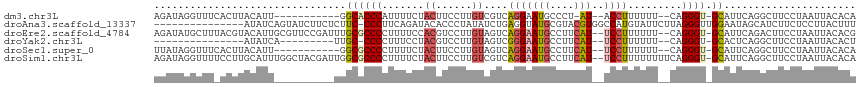
>dm3.chr3L 14546187 100 + 24543557 AGAUAGGUUUCACUUACAUU-----------GGCACCCAUUUUCUACUUCCUUGUCGUCAGGAAUGCCCU-AU--ACCUUUUUU--CAGGGU-GCAUUCAGGCUUCCUAAUUACACA .((((((............(-----------((...)))...........))))))...(((((.(((..-.(--(((((....--.)))))-)......))))))))......... ( -18.50, z-score = -0.44, R) >droAna3.scaffold_13337 18747913 101 - 23293914 ---------------AUAUCAGUAUCUUCUCUUC-CCCUUCAGAUACACCCUAUAUCUGAGGUAUGCGUACGUGGCCAUGUAUUCUUAGGGUUGGAAUAGCAUCUUCUCCUUACUUU ---------------......((...((((...(-(((((((((((.......)))))))))...(.((((((....)))))).)...)))..))))..))................ ( -19.40, z-score = -0.04, R) >droEre2.scaffold_4784 14515579 112 + 25762168 AGAUAUGCUUUACGUACAUUGCGUUCCGAUUUGCGCCCCUUUUCCACGUCCUUGUAGUCAGGAAUGCCUUCAU--UCCUUUUUU--CAGGGU-GCAUUCAGACUUCCUAAUUACACG .(..((((............))))..)((..(((((((.......(((....)))....(((((((....)))--)))).....--..))))-))).)).................. ( -22.80, z-score = -1.53, R) >droYak2.chr3L 14632045 87 + 24197627 ---------------AUAUCA---------UUGC-CCCCUUUCCUACGUCCUUGUAGUCGGGAAUGCCUUCAU--UCCUUUUUU--CAGGGU-GCACUCAGGCUUCCUAAUUACACU ---------------......---------.(((-.((((...(((((....)))))..(((((((....)))--)))).....--.)))).-)))...(((...)))......... ( -19.50, z-score = -1.91, R) >droSec1.super_0 6671446 101 + 21120651 UUAUAGGUUUCACUUACAUU-----------GGCGCCCCUUUUCUACUUCCUUGUAGUCAGGAAUGCCUUCAU--UCCUUUUUU--CAGGGU-GCAUUCAGGCUUCCUAAUUACACA ...((((.....((......-----------.((((((.....((((......))))..(((((((....)))--)))).....--..))))-))....))....))))........ ( -24.50, z-score = -2.77, R) >droSim1.chr3L 13871859 114 + 22553184 AGAUAGGUUUUCCUUGCAUUUGGCUACGAUUGGCGCCCCUUUUCUACUUCCUUGUCGUCAGGAAUGCCUUCAU--UCCUUUUUUUUCAGGGU-GCAUUCAGGCUUCCUAAUUACACA ...((((....(((.((.....))...((...((((((.......((......))....(((((((....)))--)))).........))))-))..)))))...))))........ ( -25.20, z-score = -0.61, R) >consensus AGAUAGGUUU_ACUUACAUU__________UGGCGCCCCUUUUCUACUUCCUUGUAGUCAGGAAUGCCUUCAU__UCCUUUUUU__CAGGGU_GCAUUCAGGCUUCCUAAUUACACA ....................................................(((.((.(((((.((((......((((........))))........))))))))).)).))).. ( -8.74 = -9.39 + 0.65)
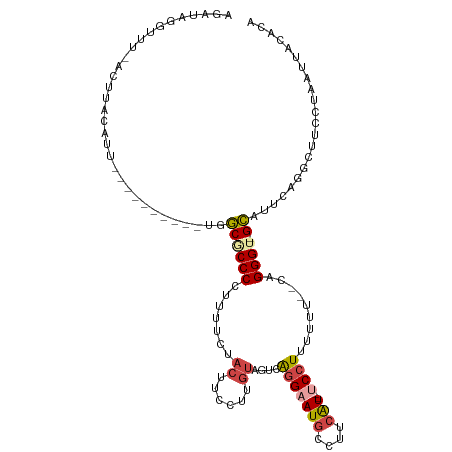
| Location | 14,546,187 – 14,546,287 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.41 |
| Shannon entropy | 0.63615 |
| G+C content | 0.43275 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.89 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14546187 100 - 24543557 UGUGUAAUUAGGAAGCCUGAAUGC-ACCCUG--AAAAAAGGU--AU-AGGGCAUUCCUGACGACAAGGAAGUAGAAAAUGGGUGCC-----------AAUGUAAGUGAAACCUAUCU ..(((..(((((((((((..(((.-..(((.--.....))))--))-.)))).)))))))..)))............((((((..(-----------(.......))..)))))).. ( -24.00, z-score = -1.36, R) >droAna3.scaffold_13337 18747913 101 + 23293914 AAAGUAAGGAGAAGAUGCUAUUCCAACCCUAAGAAUACAUGGCCACGUACGCAUACCUCAGAUAUAGGGUGUAUCUGAAGGG-GAAGAGAAGAUACUGAUAU--------------- ..((((.((((.((...)).))))..((((..(..(((........)))..).....((((((((.....))))))))))))-..........)))).....--------------- ( -20.30, z-score = -0.24, R) >droEre2.scaffold_4784 14515579 112 - 25762168 CGUGUAAUUAGGAAGUCUGAAUGC-ACCCUG--AAAAAAGGA--AUGAAGGCAUUCCUGACUACAAGGACGUGGAAAAGGGGCGCAAAUCGGAACGCAAUGUACGUAAAGCAUAUCU ...(((.....(...(((((.(((-.((((.--.....((((--(((....)))))))..((((......))))....)))).)))..)))))...)....)))............. ( -29.30, z-score = -1.91, R) >droYak2.chr3L 14632045 87 - 24197627 AGUGUAAUUAGGAAGCCUGAGUGC-ACCCUG--AAAAAAGGA--AUGAAGGCAUUCCCGACUACAAGGACGUAGGAAAGGGG-GCAA---------UGAUAU--------------- .(((((.(((((...))))).)))-))(((.--.....))).--......((...(((..((((......))))....))).-))..---------......--------------- ( -22.90, z-score = -1.79, R) >droSec1.super_0 6671446 101 - 21120651 UGUGUAAUUAGGAAGCCUGAAUGC-ACCCUG--AAAAAAGGA--AUGAAGGCAUUCCUGACUACAAGGAAGUAGAAAAGGGGCGCC-----------AAUGUAAGUGAAACCUAUAA ..((((.(((((((((((..((..-..(((.--.....))).--))..)))).))))))).))))............(((..(((.-----------.......)))...))).... ( -26.50, z-score = -2.47, R) >droSim1.chr3L 13871859 114 - 22553184 UGUGUAAUUAGGAAGCCUGAAUGC-ACCCUGAAAAAAAAGGA--AUGAAGGCAUUCCUGACGACAAGGAAGUAGAAAAGGGGCGCCAAUCGUAGCCAAAUGCAAGGAAAACCUAUCU .(((((.(((((...))))).)))-))(((........((((--(((....))))))).......)))..(((.......(((..........)))...))).(((....))).... ( -26.86, z-score = -0.95, R) >consensus UGUGUAAUUAGGAAGCCUGAAUGC_ACCCUG__AAAAAAGGA__AUGAAGGCAUUCCUGACUACAAGGAAGUAGAAAAGGGGCGCAA__________AAUGUAAGU_AAACCUAUCU ..(((..(((((((((((.........(((........))).......)))).)))))))..))).................................................... (-12.69 = -12.89 + 0.20)
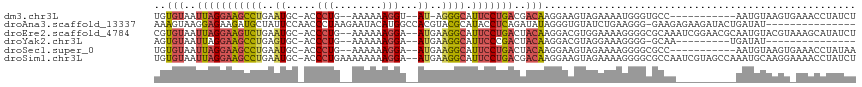
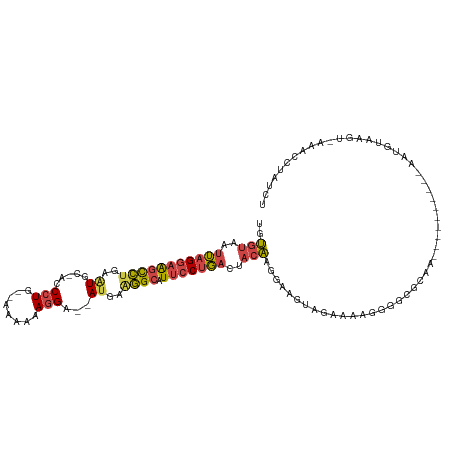
| Location | 14,546,213 – 14,546,317 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.21 |
| Shannon entropy | 0.46170 |
| G+C content | 0.39543 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.89 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14546213 104 - 24543557 ----------UAAUAGCAUAUAAGAGCAAAAAAAGAGAGUUGUGUAAUUAGGAAGCCUGAAUGC-ACCCUGA--AAAAAGGU--AU-AGGGCAUUCCUGACGACAAGGAAGUAGAAAAUG ----------.....((........))...............(((..(((((((((((..(((.-..(((..--....))))--))-.)))).)))))))..)))............... ( -19.90, z-score = -1.79, R) >droAna3.scaffold_13337 18747934 119 + 23293914 AGAGAUUUGACAAUCUCUUCUCGGAGA-GACGAAAAGGUAAAAGUAAGGAGAAGAUGCUAUUCCAACCCUAAGAAUACAUGGCCACGUACGCAUACCUCAGAUAUAGGGUGUAUCUGAAG (((((((....)))))))..(((....-..)))...((((...(((.(..(...(((.(((((.........))))))))..)..).)))...))))((((((((.....)))))))).. ( -25.10, z-score = -0.08, R) >droEre2.scaffold_4784 14515616 102 - 25762168 ----------UAAUAUC--AUAAGAGC-AACAAAGAGAGUCGUGUAAUUAGGAAGUCUGAAUGC-ACCCUGA--AAAAAGGA--AUGAAGGCAUUCCUGACUACAAGGACGUGGAAAAGG ----------.......--........-..........(((.((((.(((((((((((..((..-..(((..--....))).--))..)))).))))))).))))..))).......... ( -23.10, z-score = -2.08, R) >droYak2.chr3L 14632057 104 - 24197627 ----------UAAUAUCGUAUAAGAGC-AAAAAAGAGAGAAGUGUAAUUAGGAAGCCUGAGUGC-ACCCUGA--AAAAAGGA--AUGAAGGCAUUCCCGACUACAAGGACGUAGGAAAGG ----------.....((.(((......-.............(((((.(((((...))))).)))-))(((..--.....(((--(((....))))))........)))..))).)).... ( -20.96, z-score = -1.84, R) >droSec1.super_0 6671472 104 - 21120651 ----------UAAUGGCAUAUAAGAGC-AAAAAAGAGAGUUGUGUAAUUAGGAAGCCUGAAUGC-ACCCUGA--AAAAAGGA--AUGAAGGCAUUCCUGACUACAAGGAAGUAGAAAAGG ----------.....((........))-.............(((((.(((((...))))).)))-))(((..--....((((--(((....)))))))..((((......))))...))) ( -22.90, z-score = -2.53, R) >droSim1.chr3L 13871896 106 - 22553184 ----------UAAUAGCAUAUAAGAGC-AAAAAAGAGAGUUGUGUAAUUAGGAAGCCUGAAUGC-ACCCUGAAAAAAAAGGA--AUGAAGGCAUUCCUGACGACAAGGAAGUAGAAAAGG ----------.....((........))-.............(((((.(((((...))))).)))-))(((........((((--(((....))))))).......)))............ ( -21.86, z-score = -2.94, R) >consensus __________UAAUAGCAUAUAAGAGC_AAAAAAGAGAGUUGUGUAAUUAGGAAGCCUGAAUGC_ACCCUGA__AAAAAGGA__AUGAAGGCAUUCCUGACUACAAGGAAGUAGAAAAGG ..........................................(((..(((((((((((.........(((........))).......)))).)))))))..)))............... (-12.69 = -12.89 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:05 2011