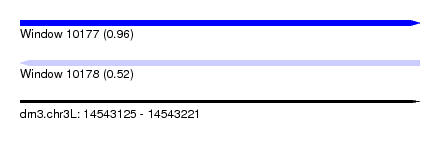
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,543,125 – 14,543,221 |
| Length | 96 |
| Max. P | 0.961464 |

| Location | 14,543,125 – 14,543,221 |
|---|---|
| Length | 96 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.90 |
| Shannon entropy | 0.63174 |
| G+C content | 0.44185 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -12.57 |
| Energy contribution | -13.04 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14543125 96 + 24543557 UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGCGAAAUCAU--UUU-AUAUA-UGGCUUCGAUCUGC--------GCUGAUUACA ...((((((((.........((((.....))))........-----...))))))))(((((((..(((--...-....)-))..))).....))--------))........ ( -26.05, z-score = -1.58, R) >droSim1.chr3L 13868871 96 + 22553184 UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGCGAAAUCAU--UUU-AUAUA-UGGCUUCGAUCUGC--------GCUGAUUACA ...((((((((.........((((.....))))........-----...))))))))(((((((..(((--...-....)-))..))).....))--------))........ ( -26.05, z-score = -1.58, R) >droSec1.super_0 6668365 96 + 21120651 UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGCGAAAUCAU--UUU-AUAUA-UGGCUUCGAUCUGC--------GCUGAUUACA ...((((((((.........((((.....))))........-----...))))))))(((((((..(((--...-....)-))..))).....))--------))........ ( -26.05, z-score = -1.58, R) >droYak2.chr3L 14628732 96 + 24197627 UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGCGAAAUCAU--UUU-AUAUA-UGGCUUCGAUCUGC--------GCUGAUUACA ...((((((((.........((((.....))))........-----...))))))))(((((((..(((--...-....)-))..))).....))--------))........ ( -26.05, z-score = -1.58, R) >droEre2.scaffold_4784 14509595 97 + 25762168 UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGCGAAAUCAU--UUUUAUAUA-UGGCUUCGAUCUGC--------GCUGAUUACA ...((((((((.........((((.....))))........-----...))))))))(((((((..(((--........)-))..))).....))--------))........ ( -26.35, z-score = -1.62, R) >droAna3.scaffold_13337 18744439 97 - 23293914 UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGUGAAAUCAU--UUUUAUAUU-GAUUUUCGAUAGGC--------GCUGAUAUCA ...((((((((.(........).))))))))......((.(-----((.((((..((((..(((((((.--........)-))))))..)))).)--------))).))))). ( -29.60, z-score = -2.95, R) >dp4.chrXR_group6 7239269 104 + 13314419 UGUAGUCCGCCAGAAAACUGACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGCGAAAUCAU--UUU-AUAUA-GAUCUGUGAUCUGCUGUUUCCUAUUGAUUAUA ..(((((((((.......(.((((.....)))).)......-----...))))))))).....(((((.--...-(((((-((((...)))))).))).......)))))... ( -25.89, z-score = -1.04, R) >droPer1.super_41 592308 104 + 728894 UGUAGUCCGCCAGAAAACUGACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGCGAAAUCAU--UUU-AUAUA-GAUCUGUGAUCUGCUGUUUCCUAUUGAUUAUA ..(((((((((.......(.((((.....)))).)......-----...))))))))).....(((((.--...-(((((-((((...)))))).))).......)))))... ( -25.89, z-score = -1.04, R) >droWil1.scaffold_180955 431668 80 + 2875958 UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGCGGACUGUGUGAAAUCAU--UUUUGUAUU-GAUUUUG------------------------- ...((((((((.........((((.....))))........-----...))))))))....(((((((.--........)-)))))).------------------------- ( -23.15, z-score = -2.97, R) >droVir3.scaffold_13049 16624625 86 + 25233164 UGUAGUCCGCUAGGAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGUGGACUGUGUGAAAUCAU--UUU-AUAGGCUGUCGUUUCUCUA------------------- ..(((((((((((....))....))))))))).........-----...((..(.(((((((....)))--...-)))).)..)).........------------------- ( -20.80, z-score = -0.56, R) >droMoj3.scaffold_6680 4673489 85 - 24764193 UAUAGUCCGCUAGGAAACUAACGGGUGGACUGUGACUGAAU-----AUGGGUGGACUGUGCGAAAUCAU--UUU-AUAGA-UGGAUUUUCUCUA------------------- (((((((((((((....)).((((.....))))........-----...))))))))))).((((((((--(..-...))-)))..))))....------------------- ( -23.60, z-score = -2.37, R) >droGri2.scaffold_15110 8080395 92 - 24565398 UAUAGUCCACCAUGAAACUAAUGGGUGGACUGUGACUGAAU-----AUGGGUGGACUGUGUGAAAUCAU--UUU-AUAGAUUGCUUUCUUUCUGUAGGCG------------- (((((((((((((.......)).)))))))))))((.(((.-----..(((..(.(((((((....)))--...-)))).)..)))...))).)).....------------- ( -25.60, z-score = -2.24, R) >anoGam1.chrU 50918180 89 - 59568033 -----------AAAAAAUCAAACGCUUGACUUCGGCUCGAUUGGUAACGGAUGCUCUGUCGGGAAUCAC-GGUAGAUAAC-GUUGACAGAUCUAAUCUAACG----------- -----------.....(((((.((...(.(....)).)).)))))...((((..((((((((..(((..-....)))..)-..)))))))....))))....----------- ( -16.10, z-score = 0.42, R) >triCas2.chrUn_141 59436 90 + 118057 UGUAGUCGGAAAGAUAUCCUGUUGGAGGGCUGUGGCUAAAC-----ACCGGCGGACUGUGUGCCACCAACAUUUCAUAAGCUACUGCAGGACACA------------------ ((((((.(((......)))((((((..((.(((......))-----)))((((.......)))).))))))...........)))))).......------------------ ( -23.50, z-score = 0.66, R) >consensus UGAAGUCCGCCAGAAAACUAACGGGUGGACUGUGACUGAAU_____AUGGGCGGACUGUGCGAAAUCAU__UUU_AUAUA_UGGCUUCGAUCUGC__________UGAUUA_A ...((((((((((....)).((((.....))))................))))))))........................................................ (-12.57 = -13.04 + 0.47)

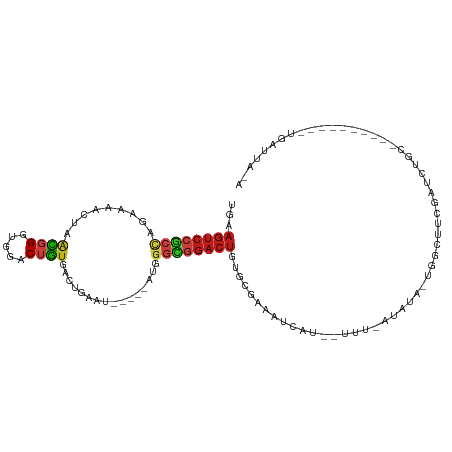
| Location | 14,543,125 – 14,543,221 |
|---|---|
| Length | 96 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.90 |
| Shannon entropy | 0.63174 |
| G+C content | 0.44185 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -7.55 |
| Energy contribution | -7.51 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.86 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
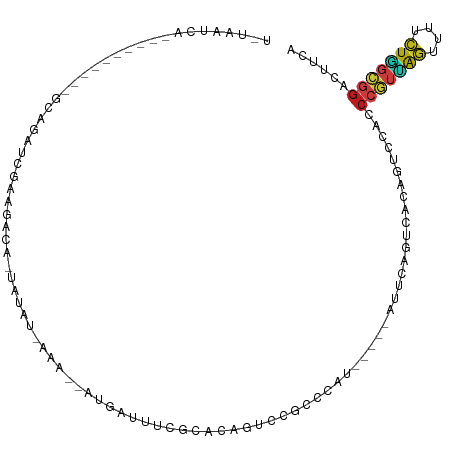
>dm3.chr3L 14543125 96 - 24543557 UGUAAUCAGC--------GCAGAUCGAAGCCA-UAUAU-AAA--AUGAUUUCGCACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA ........((--------(.((((((......-.....-...--.)))))))))..((((((((...-----..............................))))))))... ( -17.97, z-score = -0.80, R) >droSim1.chr3L 13868871 96 - 22553184 UGUAAUCAGC--------GCAGAUCGAAGCCA-UAUAU-AAA--AUGAUUUCGCACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA ........((--------(.((((((......-.....-...--.)))))))))..((((((((...-----..............................))))))))... ( -17.97, z-score = -0.80, R) >droSec1.super_0 6668365 96 - 21120651 UGUAAUCAGC--------GCAGAUCGAAGCCA-UAUAU-AAA--AUGAUUUCGCACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA ........((--------(.((((((......-.....-...--.)))))))))..((((((((...-----..............................))))))))... ( -17.97, z-score = -0.80, R) >droYak2.chr3L 14628732 96 - 24197627 UGUAAUCAGC--------GCAGAUCGAAGCCA-UAUAU-AAA--AUGAUUUCGCACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA ........((--------(.((((((......-.....-...--.)))))))))..((((((((...-----..............................))))))))... ( -17.97, z-score = -0.80, R) >droEre2.scaffold_4784 14509595 97 - 25762168 UGUAAUCAGC--------GCAGAUCGAAGCCA-UAUAUAAAA--AUGAUUUCGCACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA ........((--------(.((((((......-.........--.)))))))))..((((((((...-----..............................))))))))... ( -17.90, z-score = -0.78, R) >droAna3.scaffold_13337 18744439 97 + 23293914 UGAUAUCAGC--------GCCUAUCGAAAAUC-AAUAUAAAA--AUGAUUUCACACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA .(((((..((--------(.((.....(((((-(........--.)))))).....))..)))..))-----)))..............(((((((....)))))))...... ( -15.20, z-score = -0.60, R) >dp4.chrXR_group6 7239269 104 - 13314419 UAUAAUCAAUAGGAAACAGCAGAUCACAGAUC-UAUAU-AAA--AUGAUUUCGCACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUCAGUUUUCUGGCGGACUACA ...(((((...(....)...(((((...))))-)....-...--.)))))......((((((((...-----..............................))))))))... ( -19.41, z-score = -1.56, R) >droPer1.super_41 592308 104 - 728894 UAUAAUCAAUAGGAAACAGCAGAUCACAGAUC-UAUAU-AAA--AUGAUUUCGCACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUCAGUUUUCUGGCGGACUACA ...(((((...(....)...(((((...))))-)....-...--.)))))......((((((((...-----..............................))))))))... ( -19.41, z-score = -1.56, R) >droWil1.scaffold_180955 431668 80 - 2875958 -------------------------CAAAAUC-AAUACAAAA--AUGAUUUCACACAGUCCGCCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA -------------------------..(((((-(........--.)))))).....((((((((...-----..............................))))))))... ( -13.21, z-score = -2.04, R) >droVir3.scaffold_13049 16624625 86 - 25233164 -------------------UAGAGAAACGACAGCCUAU-AAA--AUGAUUUCACACAGUCCACCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUCCUAGCGGACUACA -------------------(((......(((.......-...--..((((......)))).......-----.....))).........(((((((....))))))).))).. ( -11.65, z-score = -0.89, R) >droMoj3.scaffold_6680 4673489 85 + 24764193 -------------------UAGAGAAAAUCCA-UCUAU-AAA--AUGAUUUCGCACAGUCCACCCAU-----AUUCAGUCACAGUCCACCCGUUAGUUUCCUAGCGGACUAUA -------------------((((.........-)))).-...--............(((((......-----.....((........))..(((((....))))))))))... ( -10.10, z-score = -0.64, R) >droGri2.scaffold_15110 8080395 92 + 24565398 -------------CGCCUACAGAAAGAAAGCAAUCUAU-AAA--AUGAUUUCACACAGUCCACCCAU-----AUUCAGUCACAGUCCACCCAUUAGUUUCAUGGUGGACUAUA -------------........((..(((.(.((((...-...--..)))).).....(......)..-----.)))..))..((((((((............))))))))... ( -14.10, z-score = -0.89, R) >anoGam1.chrU 50918180 89 + 59568033 -----------CGUUAGAUUAGAUCUGUCAAC-GUUAUCUACC-GUGAUUCCCGACAGAGCAUCCGUUACCAAUCGAGCCGAAGUCAAGCGUUUGAUUUUUU----------- -----------.....(((....((((((...-(..(((....-..)))..).))))))..))).......(((((((((........).))))))))....----------- ( -16.00, z-score = -0.33, R) >triCas2.chrUn_141 59436 90 - 118057 ------------------UGUGUCCUGCAGUAGCUUAUGAAAUGUUGGUGGCACACAGUCCGCCGGU-----GUUUAGCCACAGCCCUCCAACAGGAUAUCUUUCCGACUACA ------------------.(((((..((.((.(((....(((..((((((((.....).))))))).-----.)))))).)).)).........(((......)))))).)). ( -23.50, z-score = -0.12, R) >consensus U_UAAUCA__________GCAGAUCGAAGACA_UAUAU_AAA__AUGAUUUCGCACAGUCCGCCCAU_____AUUCAGUCACAGUCCACCCGUUAGUUUUCUGGCGGACUUCA .........................................................................................(((((((....)))))))...... ( -7.55 = -7.51 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:02 2011