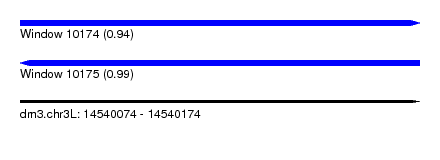
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,540,074 – 14,540,174 |
| Length | 100 |
| Max. P | 0.994377 |

| Location | 14,540,074 – 14,540,174 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.34431 |
| G+C content | 0.51796 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.06 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
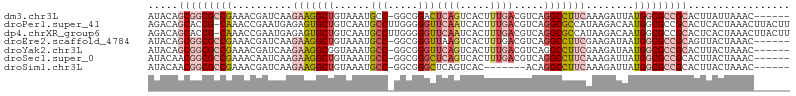
>dm3.chr3L 14540074 100 + 24543557 ------GUUUAAUAAGUGCGGCGCCAUAAUCUUUGAAGGCCUGACGUCAAAGUGACUGAGUCCGCC-GGCAUUUACAGCCUUCUUGAUCGUUUCGGCGCCGCUGUAU ------...........(((((((((..(((...((((((.((..(((.....)))...(((....-))).....))))))))..)))..)...))))))))..... ( -34.80, z-score = -2.39, R) >droPer1.super_41 589524 106 + 728894 AAGUAAGUUUAGUGAGUGCGGCGCCAUUGUCUUAUGGCGCCUGACGUCAAAGUGAUUGAACCCCCAAGGCAUUGACAGCACUCUCAUUCGGUUUG-CGGUGCUGUCU .((((......(.((((((((((((((......))))))))....(((((.((..(((......))).)).))))).)))))).)..(((.....-))))))).... ( -37.40, z-score = -2.08, R) >dp4.chrXR_group6 7236483 106 + 13314419 AAGUAAGUUUAGUGAGUGCGGCGCCAUUGUCUUAUGGCGCCUGACGUCAAAGUGAUUGAACCCCCAAGGCAUUGACAGCACUCUCAUUCGGUUUG-CGGUGCUGUCU .((((......(.((((((((((((((......))))))))....(((((.((..(((......))).)).))))).)))))).)..(((.....-))))))).... ( -37.40, z-score = -2.08, R) >droEre2.scaffold_4784 14506643 100 + 25762168 ------GUUUAGUAACUGCGGCGCCAUUAUCUUCGAAGGCCUGACGUCAAAGUGACUUAACCCGCC-GGCAUUUACAGCCUUCUUGAUCGUUUCGGCGCCGCUGUAU ------........((.((((((((.........((((((.((..(((.....)))....((....-))......))))))))..((.....)))))))))).)).. ( -33.70, z-score = -2.50, R) >droYak2.chr3L 14625700 100 + 24197627 ------GUUUAGUAAGUGCGGCGCCAUUAUCUUCGAAGGCCUGACGUCAAAGUGACUGAACCCGCC-GGCAUUUACCGCCUUCUUGAUCGUUUCGGCGCCGCUGUAU ------.....(((...((((((((.........((((((.....(((.....)))..........-((......))))))))..((.....))))))))))..))) ( -33.30, z-score = -1.72, R) >droSec1.super_0 6665413 100 + 21120651 ------GUUUAGUAAGUGCGGCGCCAUAAUCUUUGAAGGCCUGACGUCAAAGUGACUGAGCCCGCC-GGCAUUUACAGCCUUCUUGAUUGUUUCGGCGCCGUUGUAU ------.....(((...((((((((((((((...((((((.((..(((.....)))...(((....-))).....))))))))..))))))...))))))))..))) ( -39.20, z-score = -3.77, R) >droSim1.chr3L 13863863 93 + 22553184 ------GUUUAGUAAGUGCGGCGCCAUAAUCUUUGAAGGCCUGU-------GUGACUGAGCCCGCC-GGCAUUUACAGCCUUCUUGAUCGUUUCGGCGCCGUUGUAU ------.....(((...(((((((((..(((...((((((.(((-------(((.(((.(....))-)))))..)))))))))..)))..)...))))))))..))) ( -34.10, z-score = -2.56, R) >consensus ______GUUUAGUAAGUGCGGCGCCAUUAUCUUUGAAGGCCUGACGUCAAAGUGACUGAACCCGCC_GGCAUUUACAGCCUUCUUGAUCGUUUCGGCGCCGCUGUAU .................((((((((.........((((((.((..(((.....))).....((....))......))))))))..((.....))))))))))..... (-20.64 = -21.06 + 0.41)

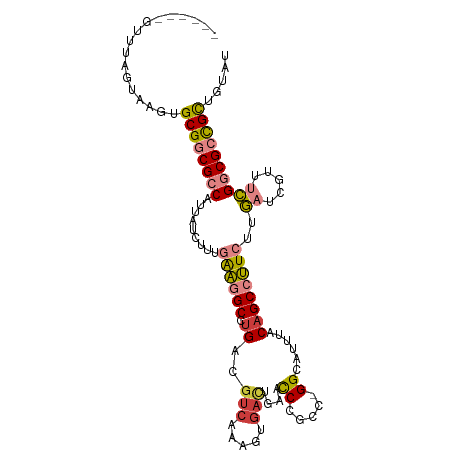
| Location | 14,540,074 – 14,540,174 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Shannon entropy | 0.34431 |
| G+C content | 0.51796 |
| Mean single sequence MFE | -36.39 |
| Consensus MFE | -20.92 |
| Energy contribution | -22.21 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.994377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14540074 100 - 24543557 AUACAGCGGCGCCGAAACGAUCAAGAAGGCUGUAAAUGCC-GGCGGACUCAGUCACUUUGACGUCAGGCCUUCAAAGAUUAUGGCGCCGCACUUAUUAAAC------ .....(((((((((....((((.....(((.......)))-(((.(((((((.....)))).)))..)))......)))).)))))))))...........------ ( -37.90, z-score = -3.94, R) >droPer1.super_41 589524 106 - 728894 AGACAGCACCG-CAAACCGAAUGAGAGUGCUGUCAAUGCCUUGGGGGUUCAAUCACUUUGACGUCAGGCGCCAUAAGACAAUGGCGCCGCACUCACUAAACUUACUU .(((((((((.-((.......)).).))))))))......((((((((((((.....)))).....((((((((......))))))))..)))).))))........ ( -36.60, z-score = -2.99, R) >dp4.chrXR_group6 7236483 106 - 13314419 AGACAGCACCG-CAAACCGAAUGAGAGUGCUGUCAAUGCCUUGGGGGUUCAAUCACUUUGACGUCAGGCGCCAUAAGACAAUGGCGCCGCACUCACUAAACUUACUU .(((((((((.-((.......)).).))))))))......((((((((((((.....)))).....((((((((......))))))))..)))).))))........ ( -36.60, z-score = -2.99, R) >droEre2.scaffold_4784 14506643 100 - 25762168 AUACAGCGGCGCCGAAACGAUCAAGAAGGCUGUAAAUGCC-GGCGGGUUAAGUCACUUUGACGUCAGGCCUUCGAAGAUAAUGGCGCCGCAGUUACUAAAC------ .....(((((((((.....(((..(((((((......(((-....)))...(((.....)))....)))))))...)))..)))))))))...........------ ( -37.60, z-score = -3.07, R) >droYak2.chr3L 14625700 100 - 24197627 AUACAGCGGCGCCGAAACGAUCAAGAAGGCGGUAAAUGCC-GGCGGGUUCAGUCACUUUGACGUCAGGCCUUCGAAGAUAAUGGCGCCGCACUUACUAAAC------ .....(((((((((.....(((..(((((((((....)))-((((...((((.....))))))))..))))))...)))..)))))))))...........------ ( -40.40, z-score = -3.80, R) >droSec1.super_0 6665413 100 - 21120651 AUACAACGGCGCCGAAACAAUCAAGAAGGCUGUAAAUGCC-GGCGGGCUCAGUCACUUUGACGUCAGGCCUUCAAAGAUUAUGGCGCCGCACUUACUAAAC------ ......((((((((....((((..(((((((......(((-....)))...(((.....)))....)))))))...)))).))))))))............------ ( -34.60, z-score = -3.19, R) >droSim1.chr3L 13863863 93 - 22553184 AUACAACGGCGCCGAAACGAUCAAGAAGGCUGUAAAUGCC-GGCGGGCUCAGUCAC-------ACAGGCCUUCAAAGAUUAUGGCGCCGCACUUACUAAAC------ ......((((((((....((((..(((((((((....(((-....)))........-------))).))))))...)))).))))))))............------ ( -31.00, z-score = -2.65, R) >consensus AUACAGCGGCGCCGAAACGAUCAAGAAGGCUGUAAAUGCC_GGCGGGUUCAGUCACUUUGACGUCAGGCCUUCAAAGAUAAUGGCGCCGCACUUACUAAAC______ .....(((((((((..........(((((((......(((.....)))((((.....)))).....)))))))........)))))))))................. (-20.92 = -22.21 + 1.29)
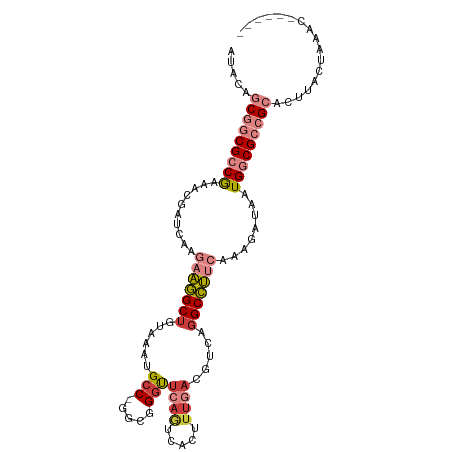
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:26:00 2011