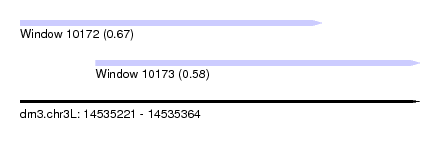
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,535,221 – 14,535,364 |
| Length | 143 |
| Max. P | 0.666814 |

| Location | 14,535,221 – 14,535,329 |
|---|---|
| Length | 108 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.69 |
| Shannon entropy | 0.63003 |
| G+C content | 0.51719 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -16.32 |
| Energy contribution | -15.22 |
| Covariance contribution | -1.09 |
| Combinations/Pair | 2.15 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14535221 108 + 24543557 CCAAUAUGGCGUAUGUGGCUCCUGGU---GCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACC .......((((...((.((..((.(.---((((((......((((...((((((((((........))))))))))))))......)))))).)---.))..)).))..)).)) ( -43.00, z-score = -1.88, R) >droGri2.scaffold_15110 8071946 105 - 24565398 CCAAUAUGGUGUAGUUGACUC---------CGAGCUAGUCUGCUUCUGCGCCUGGGCAAGUGCAUCCGUCAGGGUGGACCGCAUCUGUUGGCUGUUGGAGCUGCGAUUCUGUGA ....(((((....((.(.(((---------(((..(((((.((...(((((....))..((.(((((....))))).)).)))...)).)))))))))))).))....))))). ( -33.20, z-score = 0.78, R) >droMoj3.scaffold_6680 4665692 105 - 24764193 CCAAUAUGGCGUAUUUGGUUCCUGGG---UCCAGUUGUUCUGUUGU---GCUUUGGCAAGUGCAUCGGUUAAAGUGGAUCGCAUUUGCUGGCUG---GAACUACGAUUUUGACC .(((.....(((....((((((..((---((((.((...(((.((.---.(((....)))..)).)))...)).))))))((....)).....)---))))))))...)))... ( -30.60, z-score = -0.84, R) >droVir3.scaffold_13049 16616890 108 + 25233164 CCAAUAUGGUGUAUUGGACUCUUGAG---UCCCGUUAGUCUGCUGUUGGACUUUGGCAAGUGCAUCGGUUAAGGUGGAUCGCAUUUGCUGGCUG---GAGUUACGGGUUUGACC (((((((...)))))))((((.(((.---(((.((((((((......)))))..((((((((((((.(......).))).)))))))))))).)---)).))).))))...... ( -34.60, z-score = -1.27, R) >droWil1.scaffold_180955 422334 111 + 2875958 CCAAUAUGGUGUAUUUGUAUCACCGGCACUCCAACCAUUCGAUUCUUGAGCCUUGGACAAAGCAUCGGUUAAGGUUGAACGCAUUUGUUGACUG---GAGUUACGAUUUUGUCC .......((((.........))))((((...(((((..(((((.(((...(....)...))).)))))....))))).......((((.(((..---..)))))))...)))). ( -22.20, z-score = 1.22, R) >droPer1.super_41 584367 108 + 728894 CCAAUAUGGUGUAAGCGGCUCUUGAU---UCCAGCCGUUUUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUGACUG---GAGUUGCGAUUCUGACC ((.....)).....((((((((....---..((((......(((((...(((((((((........))))))))))))))......))))...)---))))))).......... ( -39.10, z-score = -0.90, R) >dp4.chrXR_group6 7231357 108 + 13314419 CCAAUAUGGUGUAAGCGGCUCUUGAU---UCCAGCCGUUUUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUGACUG---GAGUUGCGAUUCUGACC ((.....)).....((((((((....---..((((......(((((...(((((((((........))))))))))))))......))))...)---))))))).......... ( -39.10, z-score = -0.90, R) >droAna3.scaffold_13337 18736711 108 - 23293914 CCAAUAGGGGGUAUGGGGCUCCUGGU---UCCAGCUGUUCUGGUCCUGAACCUUGGCCAUGGCAUCGGUCAGGGUGGACCUCAUUUGCUGGCUA---GAGUUGCGAUUCUGACC .......(((((.((.(((((.(((.---.(((((......(((((...(((((((((........))))))))))))))......))))))))---))))).))))))).... ( -42.70, z-score = -0.78, R) >droEre2.scaffold_4784 14501785 108 + 25762168 CCAAUAUGGCGUAUGUGGCUCCUGGU---GCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACC .......((((...((.((..((.(.---((((((......((((...((((((((((........))))))))))))))......)))))).)---.))..)).))..)).)) ( -43.00, z-score = -1.88, R) >droYak2.chr3L 14620946 108 + 24197627 CCAAUACGGCGUAUGUGGCUCCUGGU---GCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACC ......(((.((.((..((((...(.---((((((......((((...((((((((((........))))))))))))))......)))))).)---))))..)))).)))... ( -44.90, z-score = -2.42, R) >droSec1.super_0 6660642 108 + 21120651 CCAAUAUGGCGUAUGUGGCUCCUGGU---GCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCUUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACC .......((((...((.((..((.(.---((((((......((((...((((((((((........))))))))))))))......)))))).)---.))..)).))..)).)) ( -43.00, z-score = -2.02, R) >droSim1.chr3L 13858922 108 + 22553184 CCAAUAUGGCGUAUGUGGCUCCUGGU---GCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACC .......((((...((.((..((.(.---((((((......((((...((((((((((........))))))))))))))......)))))).)---.))..)).))..)).)) ( -43.00, z-score = -1.88, R) >triCas2.chrUn_141 58491 92 + 118057 -------------UUUGUUUA---------AAAACAAAAUUAUUGUCGUUUUUUACAAAGCGCUCUUCGCAAAAAUGUUUUUUCCUCCAUUUUAAUCAGCAUGUAAGAUUCAAU -------------((((((..---------..)))))).........(..(((((((..((((.....)).((((((..........)))))).....)).)))))))..)... ( -12.00, z-score = -1.07, R) >consensus CCAAUAUGGCGUAUGUGGCUCCUGGU___UCCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUCAUUUGCUGGCUG___GAGUUGCGAUUCUGACC ..............((((((............))))))...((((....(((((((((........))))))))).........((((.((((.....))))))))....)))) (-16.32 = -15.22 + -1.09)

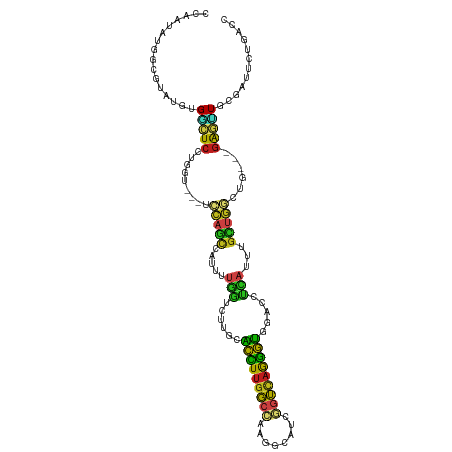
| Location | 14,535,248 – 14,535,364 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Shannon entropy | 0.55559 |
| G+C content | 0.50538 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -19.22 |
| Energy contribution | -18.77 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.77 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
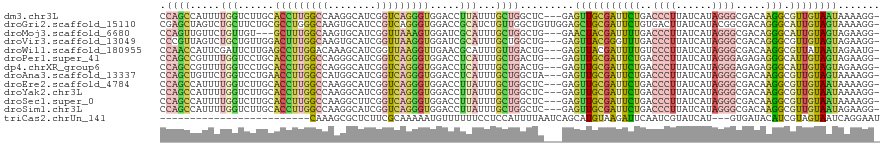
>dm3.chr3L 14535248 116 + 24543557 CCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACCCUUAUCAUAGGGCGACAAGGCGUUGUAAUAAAAGG- (((((......((((...((((((((((........))))))))))))))......)))))...---..((((((((.((..((((......))))......)).))))))))......- ( -41.20, z-score = -1.69, R) >droGri2.scaffold_15110 8071967 119 - 24565398 CGAGCUAGUCUGCUUCUGCGCCUGGGCAAGUGCAUCCGUCAGGGUGGACCGCAUCUGUUGGCUGUUGGAGCUGCGAUUCUGUGACUUAUCAUACGGCGACAGGGCAUUGUAGUAAAAGG- .((((......))))((((((....))..((.(((((....))))).)).((.(((((((.((((..(((..(((....)))..))).....)))))))))))))...)))).......- ( -36.30, z-score = 1.29, R) >droMoj3.scaffold_6680 4665719 113 - 24764193 CCAGUUGUUCUGUUGU---GCUUUGGCAAGUGCAUCGGUUAAAGUGGAUCGCAUUUGCUGGCUG---GAACUACGAUUUUGACCCUUAUCAUAGGGCGACAGGGCAUUGUAGUAGAAGG- .((((.(((((((((.---(((..((((((((((((.(......).))).))))))))))))..---...............((((......)))))))))))))))))..........- ( -34.80, z-score = -1.08, R) >droVir3.scaffold_13049 16616917 116 + 25233164 CCCGUUAGUCUGCUGUUGGACUUUGGCAAGUGCAUCGGUUAAGGUGGAUCGCAUUUGCUGGCUG---GAGUUACGGGUUUGACCCUUAUCAUAGGGCGACAGGGCGUUGUAGUAGAAGG- .((.....((((((((..(((.(..(((((((((((.(......).))).))))))))..)(((---......))))))..))(((......)))(((((.....))))))))))).))- ( -36.30, z-score = -0.23, R) >droWil1.scaffold_180955 422364 116 + 2875958 CCAACCAUUCGAUUCUUGAGCCUUGGACAAAGCAUCGGUUAAGGUUGAACGCAUUUGUUGACUG---GAGUUACGAUUUUGUCCCUUAUCAUAGGGCGACAAGGCGUUAUAAUAGAAUG- .(((((..(((((.(((...(....)...))).)))))....)))))(((((..((((.(((..---..))))))).(((((((((((...))))).)))))))))))...........- ( -30.90, z-score = -1.44, R) >droPer1.super_41 584394 116 + 728894 CCAGCCGUUUUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUGACUG---GAGUUGCGAUUCUGACCCUUAUCAUAGGGAGAGAGGGCAUUGUAGUAGAAGG- .((((......(((((...(((((((((........))))))))))))))......))))((((---.(((.((..((((..((((......))))..)))).))))).))))......- ( -44.10, z-score = -1.22, R) >dp4.chrXR_group6 7231384 116 + 13314419 CCAGCCGUUUUGGUCCUGCACCUUGGCCAGGGCAUCGGUCAGGGUGGACCUCAUUUGCUGACUG---GAGUUGCGAUUCUGACCCUUAUCAUAGGGAGAGAGGGCAUUGUAGUAGAAGG- .((((......(((((...(((((((((........))))))))))))))......))))((((---.(((.((..((((..((((......))))..)))).))))).))))......- ( -44.10, z-score = -1.22, R) >droAna3.scaffold_13337 18736738 116 - 23293914 CCAGCUGUUCUGGUCCUGAACCUUGGCCAUGGCAUCGGUCAGGGUGGACCUCAUUUGCUGGCUA---GAGUUGCGAUUCUGACCCUUAUCAUAGGGCGACAAGGCGUUGUAGUAAAAGG- (((((......(((((...(((((((((........))))))))))))))......))))).((---((((....))))))..((((....((.((((......)))).))....))))- ( -40.80, z-score = -0.93, R) >droEre2.scaffold_4784 14501812 116 + 25762168 CCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACCCUUAUCAUAGGGCGACAAGGCGUUGUAAUAAAAGG- (((((......((((...((((((((((........))))))))))))))......)))))...---..((((((((.((..((((......))))......)).))))))))......- ( -41.20, z-score = -1.69, R) >droYak2.chr3L 14620973 116 + 24197627 CCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACCCUUAUCAUAGGGCGACAAGGCGUUGUAAUAAAAGG- (((((......((((...((((((((((........))))))))))))))......)))))...---..((((((((.((..((((......))))......)).))))))))......- ( -41.20, z-score = -1.69, R) >droSec1.super_0 6660669 116 + 21120651 CCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCUUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACCCUUAUCAUAGGGCGACAAGGCGUUGUAAUAAAAGG- (((((......((((...((((((((((........))))))))))))))......)))))...---..((((((((.((..((((......))))......)).))))))))......- ( -41.20, z-score = -1.63, R) >droSim1.chr3L 13858949 116 + 22553184 CCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUUAUUUGCUGGCUC---GAGUUGCGAUUCUGACCCUUAUCAUAGGGCGACAAGGCGUUGUAAUAGAAGG- (((((......((((...((((((((((........))))))))))))))......))))).((---..((((((((.((..((((......))))......)).)))))))).))...- ( -41.70, z-score = -1.61, R) >triCas2.chrUn_141 58524 92 + 118057 -------------------------CAAAGCGCUCUUCGCAAAAAUGUUUUUUCCUCCAUUUUAAUCAGCAUGUAAGAUUCAAUCGUAUCAU---GUGAUACAUCGUAGUAAUCAGGAAU -------------------------....(((.....)))...........(((((............(((((...(((...)))....)))---))(((((......)).)))))))). ( -12.60, z-score = -0.32, R) >consensus CCAGCCAUUUUGGUCUUGCACCUUGGCCAAGGCAUCGGUCAGGGUGGACCUCAUUUGCUGGCUG___GAGUUGCGAUUCUGACCCUUAUCAUAGGGCGACAAGGCGUUGUAAUAAAAGG_ .((((.....(((......(((((((((........))))))))).....)))...)))).........(((((((((((..((((......)))).....))).))))))))....... (-19.22 = -18.77 + -0.45)
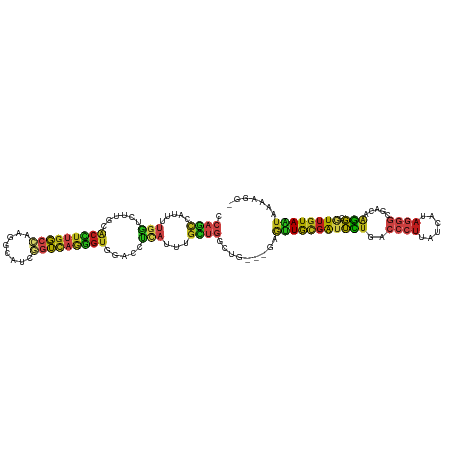
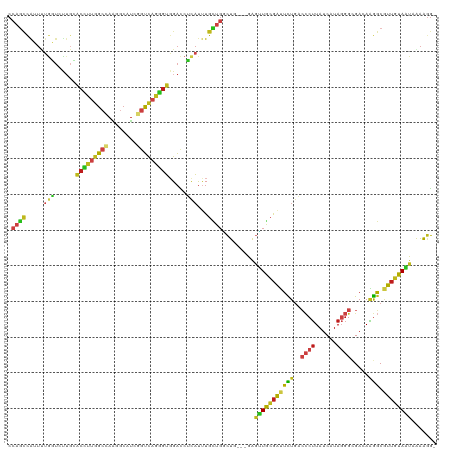
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:58 2011