| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,239,373 – 5,239,471 |
| Length | 98 |
| Max. P | 0.712522 |

| Location | 5,239,373 – 5,239,471 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Shannon entropy | 0.34588 |
| G+C content | 0.42791 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -17.78 |
| Energy contribution | -17.52 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
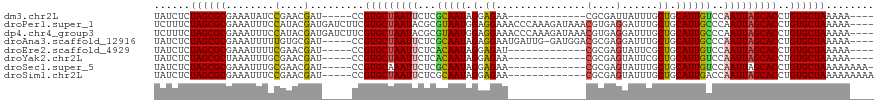
>dm3.chr2L 5239373 98 + 23011544 UAUCUCUAGCGCGAAAUAUCCGAACGAU-----CCGUGCUAAUUCUCGCAAUAGGAGAA-------------CGCGAUUAUUUGCUGCAUUGUCCAAUUAGCACCUGUGCUAAAAA---- ......((((((((((((..((.....(-----((.(((........)))...)))...-------------..))..)))))((((.(((....)))))))...)))))))....---- ( -22.90, z-score = -1.10, R) >droPer1.super_1 8480237 116 + 10282868 UCUUUCUAGCGCGAAAUUUCCAUACGAUGAUCUUCGUGCUAAUACGCGUAAUGGAGGAAACCCAAAGAUAAACGUGAGGAUUUGCUGCAUUGCCCAAUUAGCACCUGUGCUAAAAA---- ........(((((((...(((.((((...(((((((((......))))...(((.(....)))))))))...)))).)))))))).)).........((((((....))))))...---- ( -31.40, z-score = -2.43, R) >dp4.chr4_group3 7027953 116 + 11692001 UCUUUCUAGCGCGAAAUUUCCAUACGAUGAUCUUCGUGCUAAUACGCGUAAUGGAGGAAACCCAAAGAUAAACGUGAGGAUUUGCUGCAUUGCCCAAUUAGCACCUGUGCUAAAAA---- ........(((((((...(((.((((...(((((((((......))))...(((.(....)))))))))...)))).)))))))).)).........((((((....))))))...---- ( -31.40, z-score = -2.43, R) >droAna3.scaffold_12916 6255696 110 - 16180835 UAUCUCUAGCGCGAAAUUUUUGUGCGAU-----CCGUGCUAAUUCUCGCAAUAGAGGAAUGAUUG-GAUGGACGCGAGGAUUUGCUGCAUUGCCCAAUUAGCACCUGUGCUAAAAA---- ......(((((((((((((((((((.((-----(((...((.(((((......))))).))..))-))).).)))))))))))((((.(((....)))))))...)))))))....---- ( -33.30, z-score = -1.73, R) >droEre2.scaffold_4929 5317906 98 + 26641161 UAUCUCUAGCGCGAAAUUUUCGAACGAU-----CCGUGCUAAUUCUCACAAUAGGAGAU-------------CGCGAGUAUUCGCUGCAUUGUCCAAUUAGCACCUGUGCUAAAAA---- ......(((((((......((....)).-----..(((((((((...(((((.((.((.-------------(....)...)).))..)))))..))))))))).)))))))....---- ( -24.70, z-score = -1.57, R) >droYak2.chr2L 8365851 98 - 22324452 UAUCUCUAGCGCUAAAUUUGCGAACGAU-----CCGUGCUAAUUCUCACAAUAGGAGAA-------------CGCGAGUAUUCGCUGCAUUGUCCAAUUAGCACCUGUGCUAAAAA---- ......((((((.....(((....))).-----..(((((((((...((((((((((.(-------------(....)).))).))..)))))..)))))))))..))))))....---- ( -24.30, z-score = -1.43, R) >droSec1.super_5 3319880 101 + 5866729 UAUCUCUAGCGCGAAAUUUGCGAACGAU-----CCGUGCAAAUUCUCGCAAUAGGAGAA-------------CGCGAGUAUUUGCUGCAUUGUCCAAUUAGCACCUGUGCUAAAAAAAA- ........((((((((((((((.....(-----((.(((........)))...)))...-------------))))))).))))).)).........((((((....))))))......- ( -27.80, z-score = -1.90, R) >droSim1.chr2L 5037717 102 + 22036055 UAUCUCUAGCGCGAAAUUUCCGAACGAU-----CCGUGCUAAUUCUCGCAAUAGGAGAA-------------CGCGAGUAUUUGCUGCAUUGACCAAUUAGCACCUGUGCUAAAAAAAAA ......(((((((........((....)-----).(((((((((....((((((.((((-------------(....)).))).))..))))...))))))))).)))))))........ ( -23.00, z-score = -1.00, R) >consensus UAUCUCUAGCGCGAAAUUUCCGAACGAU_____CCGUGCUAAUUCUCGCAAUAGGAGAA_____________CGCGAGUAUUUGCUGCAUUGUCCAAUUAGCACCUGUGCUAAAAA____ ......((((((.............(........)(((((((((...(((((.(.((...........................)).))))))..)))))))))..))))))........ (-17.78 = -17.52 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:24 2011