| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,488,949 – 14,489,048 |
| Length | 99 |
| Max. P | 0.664235 |

| Location | 14,488,949 – 14,489,048 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Shannon entropy | 0.35465 |
| G+C content | 0.44006 |
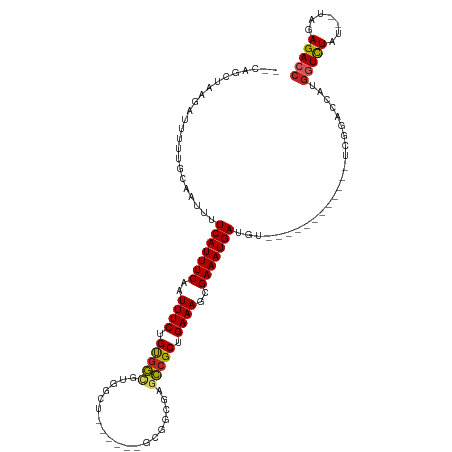
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.39 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
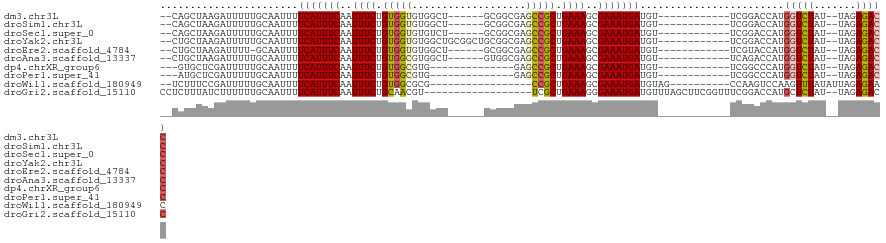
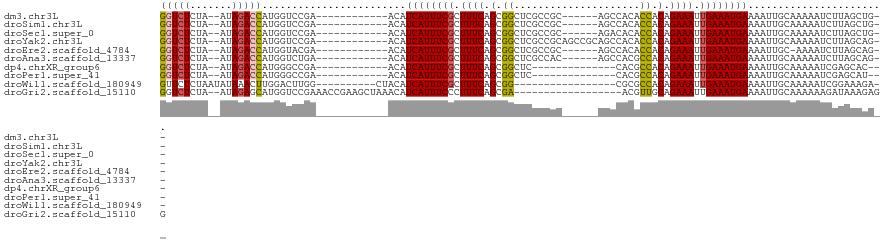
>dm3.chr3L 14488949 99 + 24543557 --CAGCUAAGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGUGUGGCU------GCGGCGAGCCGCUGAAAGCGAAAUGAUGU------------UCGGACCAUGGUCUAU--UAGAGACC --...........((((.(((..(((((((..((((.(((((...((.------...))..))))).))))..))))))).))------------)))))....(((((..--...))))) ( -26.50, z-score = -0.47, R) >droSim1.chr3L 13817772 99 + 22553184 --CAGCUAAGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGUGUGGCU------GCGGCGAGCCGCUGAAAGCGAAAUGAUGU------------UCGGACCAUGGUCUAU--UAGAGACC --...........((((.(((..(((((((..((((.(((((...((.------...))..))))).))))..))))))).))------------)))))....(((((..--...))))) ( -26.50, z-score = -0.47, R) >droSec1.super_0 6619510 99 + 21120651 --CAGCUAAGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGUGUGUCU------GCGGCGAGCCGCUGAAAGCGAAAUGAUGU------------UCGGACCAUGGUCUAU--UAGAGACC --...........((((.(((..(((((((..((((.(((((.(((..------...))).))))).))))..))))))).))------------)))))....(((((..--...))))) ( -27.10, z-score = -0.99, R) >droYak2.chr3L 14578633 105 + 24197627 --CUGCUAAGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGUGUGGCUGCGGCUGCGGCGAGCCGCUGAAAGCGAAAUGAUGU------------UCGGACCAUGGUCUAU--UAGAGACC --((.((((..............(((((((..((((.(((((...(((((....)))))..))))).))))..)))))))...------------..((((....)))).)--)))))... ( -33.00, z-score = -1.41, R) >droEre2.scaffold_4784 14460732 98 + 25762168 --CUGCUAAGAUUUU-GCAAUUUUCAUUUCAAUUUCUGUGGUGUGGCU------GCGGCGAGCCGCUGAAAGCGAAAUGAUGU------------UCGUACCAUGGUCUAU--UAGAGACC --.(((.........-)))....(((((((..((((.(((((...((.------...))..))))).))))..)))))))...------------.........(((((..--...))))) ( -26.70, z-score = -0.95, R) >droAna3.scaffold_13337 18698527 99 - 23293914 --CUGCUAAGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGCGUGGCU------GUGGCGAGCCGCUGAAAGCGAAAUGAUGU------------UCAGACCAUGGUCUAU--UAGAGACC --...........((((.(((..(((((((..((((.(((((.(.((.------...)).)))))).))))..))))))).))------------)))))....(((((..--...))))) ( -29.40, z-score = -1.62, R) >dp4.chrXR_group6 7186757 90 + 13314419 ---GUGCUCGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGCGUG--------------GAGCCGCUGAAAGCGAAAUGAUGU------------UCGGCCCAUGGUCUAU--UAGAGACC ---(.((.(((.....(((....(((((((..((((.(((((...--------------..))))).))))..))))))))))------------))))).)..(((((..--...))))) ( -28.60, z-score = -2.20, R) >droPer1.super_41 539088 90 + 728894 ---AUGCUCGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGCGUG--------------GAGCCGCUGAAAGCGAAAUGAUGU------------UCGGCCCAUGGUCUAU--UAGAGACC ---..((.(((.....(((....(((((((..((((.(((((...--------------..))))).))))..))))))))))------------)))))....(((((..--...))))) ( -28.20, z-score = -2.13, R) >droWil1.scaffold_180949 3859778 92 + 6375548 --UCUUUCCGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGCGCG-----------------CCGCUGAAAGCGAAAUGAUGUAG----------CCAAGUCCAAGUUUAUAUUAGAGAAC --.......(((((..((.((..(((((((..((((.((((....-----------------)))).))))..))))))).)).)----------).)))))................... ( -20.90, z-score = -1.21, R) >droGri2.scaffold_15110 21731410 101 + 24565398 CCUCUUUAUCUUUUUUGCAAUUUUCAUUUCAAUUUCUGCAACGU------------------UCGCUGAAAGGGAAAUGAUGUUUAGCUUCGGUUUCGGACCAUGCUCUAU--UAGAGACC ..((((((........(((....(((((((..((((.((.....------------------..)).))))..))))))).(((((((....)))..))))..))).....--)))))).. ( -22.12, z-score = -1.22, R) >consensus __CAGCUAAGAUUUUUGCAAUUUUCAUUUCAAUUUCUGUGGCGUGGCU______GCGGCGAGCCGCUGAAAGCGAAAUGAUGU____________UCGGACCAUGGUCUAU__UAGAGACC .......................(((((((..((((.(((((...(((........)))..))))).))))..)))))))........................(((((.......))))) (-19.90 = -20.39 + 0.49)
| Location | 14,488,949 – 14,489,048 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Shannon entropy | 0.35465 |
| G+C content | 0.44006 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
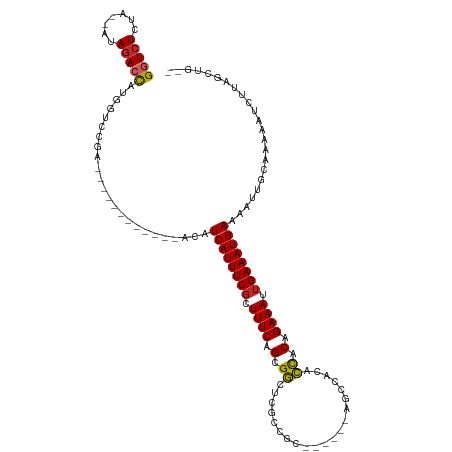
>dm3.chr3L 14488949 99 - 24543557 GGUCUCUA--AUAGACCAUGGUCCGA------------ACAUCAUUUCGCUUUCAGCGGCUCGCCGC------AGCCACACCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCUUAGCUG-- (((((...--..))))).........------------...((((((((.((((.((((....))))------.(........).)))).)))))))).....................-- ( -22.90, z-score = -1.49, R) >droSim1.chr3L 13817772 99 - 22553184 GGUCUCUA--AUAGACCAUGGUCCGA------------ACAUCAUUUCGCUUUCAGCGGCUCGCCGC------AGCCACACCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCUUAGCUG-- (((((...--..))))).........------------...((((((((.((((.((((....))))------.(........).)))).)))))))).....................-- ( -22.90, z-score = -1.49, R) >droSec1.super_0 6619510 99 - 21120651 GGUCUCUA--AUAGACCAUGGUCCGA------------ACAUCAUUUCGCUUUCAGCGGCUCGCCGC------AGACACACCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCUUAGCUG-- (((((...--..))))).........------------...((((((((.((((.((((....))))------.(......)...)))).)))))))).....................-- ( -23.50, z-score = -1.89, R) >droYak2.chr3L 14578633 105 - 24197627 GGUCUCUA--AUAGACCAUGGUCCGA------------ACAUCAUUUCGCUUUCAGCGGCUCGCCGCAGCCGCAGCCACACCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCUUAGCAG-- (((((...--..))))).........------------...((((((((.((((.((((((......)))))).(........).)))).))))))))....(((..........))).-- ( -27.30, z-score = -2.15, R) >droEre2.scaffold_4784 14460732 98 - 25762168 GGUCUCUA--AUAGACCAUGGUACGA------------ACAUCAUUUCGCUUUCAGCGGCUCGCCGC------AGCCACACCACAGAAAUUGAAAUGAAAAUUGC-AAAAUCUUAGCAG-- (((((...--..))))).........------------...((((((((.((((.((((....))))------.(........).)))).))))))))....(((-.........))).-- ( -24.00, z-score = -2.13, R) >droAna3.scaffold_13337 18698527 99 + 23293914 GGUCUCUA--AUAGACCAUGGUCUGA------------ACAUCAUUUCGCUUUCAGCGGCUCGCCAC------AGCCACGCCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCUUAGCAG-- (((((...--..))))).........------------...((((((((.((((.(.(((..((...------.))...))).).)))).))))))))....(((..........))).-- ( -23.90, z-score = -1.81, R) >dp4.chrXR_group6 7186757 90 - 13314419 GGUCUCUA--AUAGACCAUGGGCCGA------------ACAUCAUUUCGCUUUCAGCGGCUC--------------CACGCCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCGAGCAC--- (((((...--..)))))....(((((------------...((((((((.((((.(.(((..--------------...))).).)))).))))))))...((....)).))).))..--- ( -24.90, z-score = -2.66, R) >droPer1.super_41 539088 90 - 728894 GGUCUCUA--AUAGACCAUGGGCCGA------------ACAUCAUUUCGCUUUCAGCGGCUC--------------CACGCCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCGAGCAU--- (((((...--..)))))....(((((------------...((((((((.((((.(.(((..--------------...))).).)))).))))))))...((....)).))).))..--- ( -24.90, z-score = -2.48, R) >droWil1.scaffold_180949 3859778 92 - 6375548 GUUCUCUAAUAUAAACUUGGACUUGG----------CUACAUCAUUUCGCUUUCAGCGG-----------------CGCGCCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCGGAAAGA-- ((((..............))))...(----------(....((((((((.((((.(.((-----------------....)).).)))).)))))))).....))..............-- ( -16.64, z-score = -0.01, R) >droGri2.scaffold_15110 21731410 101 - 24565398 GGUCUCUA--AUAGAGCAUGGUCCGAAACCGAAGCUAAACAUCAUUUCCCUUUCAGCGA------------------ACGUUGCAGAAAUUGAAAUGAAAAUUGCAAAAAAGAUAAAGAGG ...((((.--....(((.((((.....))))..))).....(((((((..((((.((((------------------...)))).))))..)))))))..................)))). ( -19.90, z-score = -1.01, R) >consensus GGUCUCUA__AUAGACCAUGGUCCGA____________ACAUCAUUUCGCUUUCAGCGGCUCGCCGC______AGCCACACCACAGAAAUUGAAAUGAAAAUUGCAAAAAUCUUAGCUG__ (((((.......)))))........................((((((((.((((.(.((.....................)).).)))).))))))))....................... (-14.38 = -14.42 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:50 2011