| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,232,034 – 5,232,131 |
| Length | 97 |
| Max. P | 0.834993 |

| Location | 5,232,034 – 5,232,131 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.46 |
| Shannon entropy | 0.56494 |
| G+C content | 0.44995 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -11.23 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
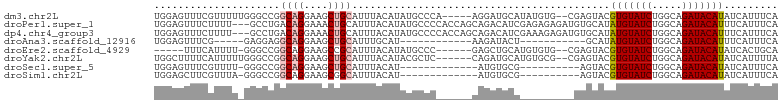
>dm3.chr2L 5232034 97 + 23011544 UGGAGUUUCGUUUUUGGGCCGGCAGGAAGCUGCAUUUACAUAUGCCCA-----AGGAUGCAUAUGUG--CGAGUACGUGUAUCUGGCAGAUACAUAUCAUUUCA .((((((((((((((((((..((((....))))..........)))))-----)))))(((....))--))))...(((((((.....)))))))...))))). ( -29.40, z-score = -1.25, R) >droPer1.super_1 8471740 101 + 10282868 UGGAGUUUCUUUU---GCCUGACAGGAAACUGCAUUUACAUAUGCCCCACCAGCAGACAUCGAGAGAGAUGUGCAUAUGUAUCUGGCAGAUACAUUUCAUUUCA ((((((....(((---(((.(((((....)))....(((((((((...........(((((......)))))))))))))))).))))))...))))))..... ( -30.90, z-score = -3.01, R) >dp4.chr4_group3 7019448 101 + 11692001 UGGAGUUUCUUUU---GCCUGACAGGAAACUGCAUUUACAUAUGCCCCACCAGCAGACAUCGAAAGAGAUGUGCAUAUGUAUCUGGCAGAUACAUUUCAUUUCA ((((((....(((---(((.(((((....)))....(((((((((.......(((....((....))..)))))))))))))).))))))...))))))..... ( -31.11, z-score = -3.29, R) >droAna3.scaffold_12916 6248763 76 - 16180835 UGGAGUUUCG-----GAGGAGGCAGGAAGCUGCAUUUGCAU------------AAGAUACU-----------GCAUAUGUAUCUGGCAGAUACAUUUCAUUUCA .........(-----((((((((((....))))((((((..------------.((((((.-----------......)))))).))))))....))).)))). ( -22.20, z-score = -2.33, R) >droEre2.scaffold_4929 5310866 90 + 26641161 -----UUUCAUUUU-GGGCCGGCAGGAAGCCGCAUUUACAUAUGCCC------GAGCUGCAUGUGUG--CGAGUACGUGUAUCUGGCAGAUACAUAUCACUGCA -----.........-......((((((.(((((....((((((((..------.....)))))))))--)).))..(((((((.....))))))).)).)))). ( -26.70, z-score = -0.03, R) >droYak2.chr2L 8358001 96 - 22324452 UGGCUUUUCAUUUUUGGGCCGGCAGGAAGCUGCAUUUACAUACGCUC------CAGAUGCAUGUGCG--CGAGUACGUGUAUCUGGCAGAUACAUAUCAUUUUA .(((((.........))))).((((....))))...........(((------(((((((((((((.--...)))))))))))))).))............... ( -38.20, z-score = -4.40, R) >droSec1.super_5 3312746 80 + 5866729 UGGAGUUUCGUUUU-GGGCCGGCAGGAAGCUGCAUUUACAU-------------AUGUGCG----------AGUACGUGUAUCUGGCAGAUACAUAUCAUUUCA .((((((((((..(-(.....((((....)))).....)).-------------....)))----------))...(((((((.....)))))))...))))). ( -20.10, z-score = -0.36, R) >droSim1.chr2L 5030500 80 + 22036055 UGGAGCUUCGUUUA-GGGCCGGCAGGAAGCGGCAUUUACAU-------------AUGUGCG----------AGUACGUGUAUCUGGCAGAUACAUAUCAUUUCA .((((((((((((.-..(....)...)))))((((......-------------..)))))----------)))..(((((((.....))))))).....))). ( -17.10, z-score = 0.76, R) >consensus UGGAGUUUCGUUUU_GGGCCGGCAGGAAGCUGCAUUUACAUAUGCCC______CAGAUGCA_________GAGUACGUGUAUCUGGCAGAUACAUAUCAUUUCA .....................((((....))))...........................................(((((((.....)))))))......... (-11.23 = -11.50 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:23 2011