| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,411,896 – 14,411,987 |
| Length | 91 |
| Max. P | 0.998886 |

| Location | 14,411,896 – 14,411,987 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 72.13 |
| Shannon entropy | 0.51871 |
| G+C content | 0.39918 |
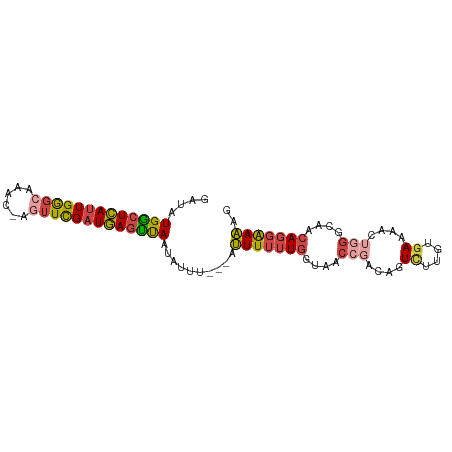
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.42 |
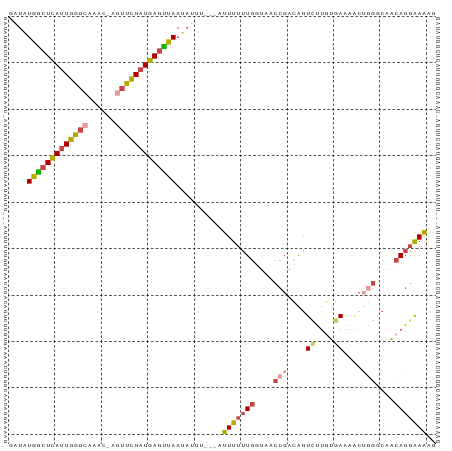
| Combinations/Pair | 1.36 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.998886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
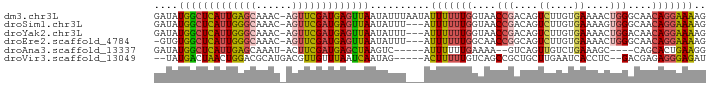
>dm3.chr3L 14411896 91 - 24543557 GAUAUGGCUCAUUGAGCAAAC-AGUUCGAUGAGUUAAUAUUUAAUAUUUUUUGGUAACCGACAGUCUUGUGAAAACUGGGCAACAGGAAAAG ....(((((((((((((....-.)))))))))))))..........(((((((((.....)).((((.((....)).))))..))))))).. ( -25.80, z-score = -3.64, R) >droSim1.chr3L 13749795 88 - 22553184 GAUAUGGCUCAUUGGGCAAAC-AGUUCGAUGAGUUAAUAUUU---AUUUUUUGGUAACCGACAGUCUUGUGAAAAGUGGGCAACAGGAAAAG ....(((((((((((((....-.)))))))))))))......---.(((((((....(((....((....))....)))....))))))).. ( -24.20, z-score = -2.96, R) >droYak2.chr3L 14505204 88 - 24197627 GAUAUGGCUCAUUGGGCAAAC-AGUUCGAUGAGUUAAUAUUU---AUUUUUUGGUAACCGACAGUCUUGUGAAAACUGGACAACAGGAAAAG ....(((((((((((((....-.)))))))))))))......---.(((((((((.....)).((((.((....)).))))..))))))).. ( -25.40, z-score = -3.57, R) >droEre2.scaffold_4784 14392419 87 - 25762168 -GUGUGGCUCAUUGGGCAAAC-AGUUCGAUGAGUUAAUAUUU---AUUUUUUGGCAACCGGCAGUCUUGUGAAAACUGGGCAACAGGAAAAG -...(((((((((((((....-.)))))))))))))......---.(((((((((.....)).((((.((....)).))))..))))))).. ( -26.70, z-score = -2.74, R) >droAna3.scaffold_13337 18632128 80 + 23293914 GAUAUGGCUCAUUGAGCAAAU-ACUUCGAUGAGCUAAGUC-----AUUUUUUGAAAA--GUCAGUUGUCUGAAAGC----CAGCACUGAAGG (((.((((((((((((.....-..)))))))))))).)))-----............--.(((((.(.(((.....----)))))))))... ( -25.30, z-score = -2.97, R) >droVir3.scaffold_13049 3314227 83 + 25233164 --UAUGACUAACUGGACGCAUGACGUUGUUUAAUCAAUAG-----ACUUUUUGUCAGCCGCUGCUUGAAUCACCUC--GACGAGAGGGAGAU --...........((..((.(((((..(((((.....)))-----))....)))))))..)).......((.((((--.....))))))... ( -16.30, z-score = 0.23, R) >consensus GAUAUGGCUCAUUGGGCAAAC_AGUUCGAUGAGUUAAUAUUU___AUUUUUUGGUAACCGACAGUCUUGUGAAAACUGGGCAACAGGAAAAG ....(((((((((((((......)))))))))))))..........(((((((....(((....((....))....)))....))))))).. (-16.69 = -17.12 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:42 2011